6S01
 
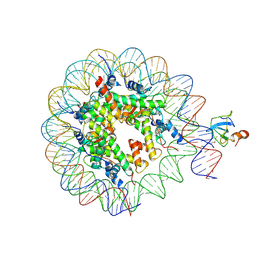 | | Structure of LEDGF PWWP domain bound H3K36 methylated nucleosome | | 分子名称: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | 著者 | Wang, H, Farnung, L, Dienemann, C, Cramer, P. | | 登録日 | 2019-06-13 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1CGM
 
 | |
8R0F
 
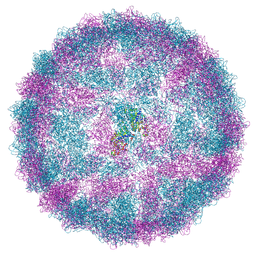 | | Capsid structure of Giardiavirus (GLV) HP strain | | 分子名称: | Capsid protein | | 著者 | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | 登録日 | 2023-10-31 | | 公開日 | 2024-04-03 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite.
Plos Pathog., 20, 2024
|
|
8R0G
 
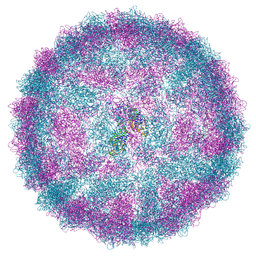 | | Capsid structure of Giardiavirus (GLV) CAT strain | | 分子名称: | Capsid protein | | 著者 | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | 登録日 | 2023-10-31 | | 公開日 | 2024-04-03 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite.
Plos Pathog., 20, 2024
|
|
1ZKL
 
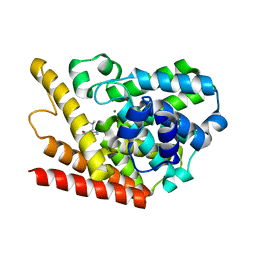 | | Multiple Determinants for Inhibitor Selectivity of Cyclic Nucleotide Phosphodiesterases | | 分子名称: | 3-ISOBUTYL-1-METHYLXANTHINE, High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | 著者 | Wang, H, Liu, Y, Chen, Y, Robinson, H, Ke, H. | | 登録日 | 2005-05-03 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7
J.Biol.Chem., 280, 2005
|
|
5C3I
 
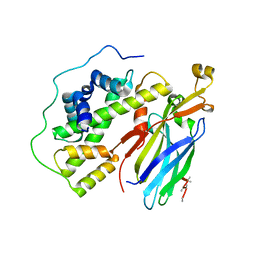 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | 分子名称: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | 著者 | Wang, H, Wang, M, Yang, N, Xu, R.M. | | 登録日 | 2015-06-17 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
7SCG
 
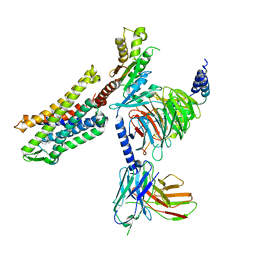 | | FH210 bound Mu Opioid Receptor-Gi Protein Complex | | 分子名称: | (2E)-N-[(2S)-2-(dimethylamino)-3-(4-hydroxyphenyl)propyl]-3-(naphthalen-1-yl)prop-2-enamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, H, Kobilka, B. | | 登録日 | 2021-09-28 | | 公開日 | 2022-04-20 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1YNS
 
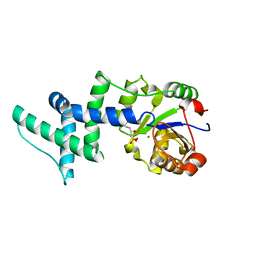 | | Crystal Structure Of Human Enolase-phosphatase E1 and its complex with a substrate analog | | 分子名称: | 2-OXOHEPTYLPHOSPHONIC ACID, E-1 enzyme, MAGNESIUM ION | | 著者 | Wang, H, Pang, H, Bartlam, M, Rao, Z. | | 登録日 | 2005-01-25 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of human e1 enzyme and its complex with a substrate analog reveals the mechanism of its phosphatase/enolase
J.Mol.Biol., 348, 2005
|
|
1RMV
 
 | | RIBGRASS MOSAIC VIRUS, FIBER DIFFRACTION | | 分子名称: | RIBGRASS MOSAIC VIRUS COAT PROTEIN, RIBGRASS MOSAIC VIRUS RNA | | 著者 | Wang, H, Stubbs, G. | | 登録日 | 1997-02-11 | | 公開日 | 1997-05-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | FIBER DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular dynamics in refinement against fiber diffraction data.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
1ZS9
 
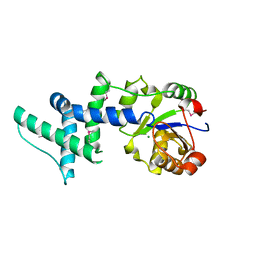 | | Crystal structure of human enolase-phosphatase E1 | | 分子名称: | E-1 ENZYME, MAGNESIUM ION | | 著者 | Wang, H, Pang, H, Bartlam, M, Rao, Z. | | 登録日 | 2005-05-23 | | 公開日 | 2005-06-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of Human E1 Enzyme and its Complex with a Substrate Analog Reveals the Mechanism of its Phosphatase/Enolase
J.Mol.Biol., 348, 2005
|
|
2NZ0
 
 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | 分子名称: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | 著者 | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | 登録日 | 2006-11-22 | | 公開日 | 2006-12-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
8EAV
 
 | | YAR027W and YAR028W in complex with c subunits from yeast VO complex | | 分子名称: | YAR027W or YAR028W, subunit from the c ring of yeast VO complex | | 著者 | Wang, H, Bueler, S.A, Rubinstein, J.L. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-02-15 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAT
 
 | | Yeast VO missing subunits a, e, and f in complex with Vma12-22p | | 分子名称: | V-type proton ATPase subunit F, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | 著者 | Wang, H, Bueler, S.A, Rubinstein, J.L. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-02 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAS
 
 | | Yeast VO in complex with Vma12-22p | | 分子名称: | V-type proton ATPase assembly factor Vma12p, V-type proton ATPase subunit F, V-type proton ATPase subunit a, ... | | 著者 | Wang, H, Bueler, S.A, Rubinstein, J.L. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-02 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAU
 
 | | Yeast VO in complex with Vma21p | | 分子名称: | V-type proton ATPase subunit a, vacuolar isoform, V-type proton ATPase subunit c, ... | | 著者 | Wang, H, Bueler, S.A, Rubinstein, J.L. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-02 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3K3E
 
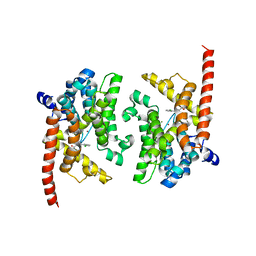 | | Crystal structure of the PDE9A catalytic domain in complex with (R)-BAY73-6691 | | 分子名称: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | 登録日 | 2009-10-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
3K3H
 
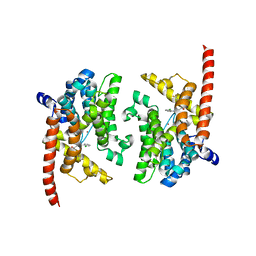 | | Crystal structure of the PDE9A catalytic domain in complex with (S)-BAY73-6691 | | 分子名称: | 1-(2-chlorophenyl)-6-[(2S)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | 登録日 | 2009-10-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
6LIV
 
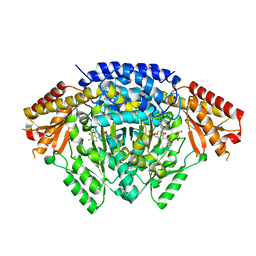 | |
6LNQ
 
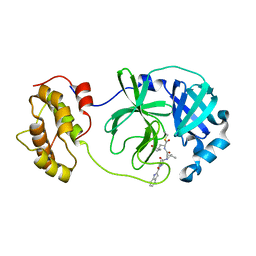 | | The co-crystal structure of SARS-CoV 3C Like Protease with aldehyde inhibitor M7 | | 分子名称: | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide, Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | | 著者 | Wang, H, Shang, L.Q. | | 登録日 | 2020-01-01 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.244 Å) | | 主引用文献 | Comprehensive Insights into the Catalytic Mechanism of Middle East Respiratory Syndrome 3C-Like Protease and Severe Acute Respiratory Syndrome 3C-Like Protease.
Acs Catalysis, 10, 2020
|
|
6LO0
 
 | |
2DY1
 
 | | Crystal structure of EF-G-2 from Thermus thermophilus | | 分子名称: | Elongation factor G, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Wang, H, Takemoto, C, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-09-04 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of EF-G-2 from Thermus thermophilus
To be Published
|
|
4MM8
 
 | |
4MMF
 
 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | 分子名称: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | 著者 | Wang, H, Gouaux, E. | | 登録日 | 2013-09-08 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MM4
 
 | |
4MM9
 
 | |
