7Y3F
 
 | | Structure of the Anabaena PSI-monomer-IsiA supercomplex | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Nagao, R, Kato, K, Hamaguchi, T, Kawakami, K, Yonekura, K, Shen, J.R. | | 登録日 | 2022-06-10 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nat Commun, 14, 2023
|
|
2FZA
 
 | | Crystal structure of d(GCGGGAGC): the base-intercalated duplex | | 分子名称: | 5'-D(*GP*(CBR)P*GP*GP*GP*AP*GP*C)-3', CALCIUM ION, SODIUM ION | | 著者 | Kondo, J, Ciengshin, T, Juan, E.C.M, Mitomi, K, Takenaka, A. | | 登録日 | 2006-02-09 | | 公開日 | 2007-01-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Crystal structure of d(gcGXGAgc) with X=G: a mutation at X is possible to occur in a base-intercalated duplex for multiplex formation
Nucleosides Nucleotides Nucleic Acids, 25, 2006
|
|
4O0L
 
 | |
1IY8
 
 | | Crystal Structure of Levodione Reductase | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, LEVODIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Sogabe, S, Fukami, T, Shiratori, Y, Yoshizumi, A, Wada, M. | | 登録日 | 2002-07-25 | | 公開日 | 2003-05-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Crystal Structure and Stereospecificity of Levodione Reductase from Corynebacterium aquaticum M-13
J.BIOL.CHEM., 278, 2003
|
|
7DC8
 
 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | 著者 | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | 登録日 | 2020-10-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.757 Å) | | 主引用文献 | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7DC7
 
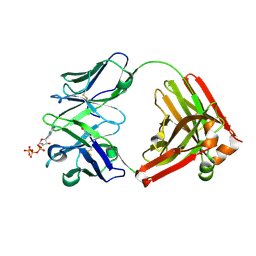 | | Crystal structure of D12 Fab-ATP complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | 著者 | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | 登録日 | 2020-10-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
1WYD
 
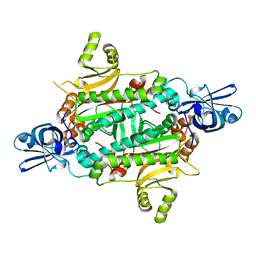 | | Crystal Structure of Aspartyl-tRNA synthetase from Sulfolobus tokodaii | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | 著者 | Maeda, Y, Hossain, M.T, Ubukata, S, Suzuki, K, Sekiguchi, T, Takenaka, A. | | 登録日 | 2005-02-12 | | 公開日 | 2006-07-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the nondiscriminating aspartyl-tRNA synthetase from the crenarchaeon Sulfolobus tokodaii strain 7 reveals the recognition mechanism for two different tRNA anticodons
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2V4C
 
 | |
2XA5
 
 | |
2WX9
 
 | |
2WYP
 
 | |
2WYK
 
 | | SiaP in complex with Neu5Gc | | 分子名称: | N-glycolyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP, THIOCYANATE ION | | 著者 | Fischer, M, Hubbard, R.E. | | 登録日 | 2009-11-16 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Water networks can determine the affinity of ligand binding to proteins.
J.Am.Chem.Soc., 2019
|
|
6H75
 
 | | SiaP A11N in complex with Neu5Ac (RT) | | 分子名称: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | 著者 | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | 登録日 | 2018-07-30 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
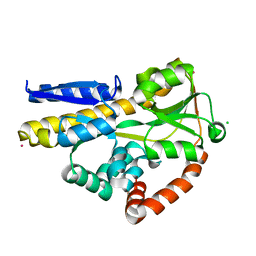 | | SiaP in complex with Neu5Ac (RT) | | 分子名称: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | 著者 | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | 登録日 | 2018-07-30 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
3A3G
 
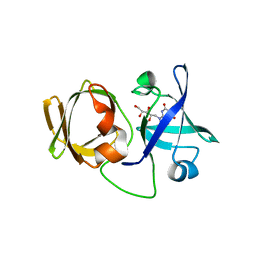 | | Crystal structure of LumP complexed with 6,7-dimethyl-8-(1'-D-ribityl) lumazine | | 分子名称: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Lumazine protein | | 著者 | Sato, Y. | | 登録日 | 2009-06-12 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3A3B
 
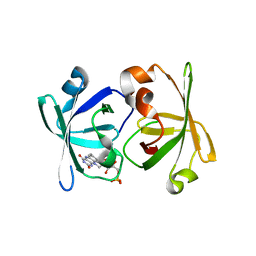 | | Crystal structure of LumP complexed with flavin mononucleotide | | 分子名称: | FLAVIN MONONUCLEOTIDE, Lumazine protein, RIBOFLAVIN | | 著者 | Sato, Y. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3A35
 
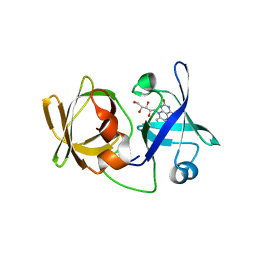 | | Crystal structure of LumP complexed with riboflavin | | 分子名称: | Lumazine protein, RIBOFLAVIN | | 著者 | Sato, Y. | | 登録日 | 2009-06-09 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.421 Å) | | 主引用文献 | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
5Y3T
 
 | | Crystal structure of hetero-trimeric core of LUBAC: HOIP double-UBA complexed with HOIL-1L UBL and SHARPIN UBL | | 分子名称: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, RanBP-type and C3HC4-type zinc finger-containing protein 1, ... | | 著者 | Tokunaga, A, Fujita, H, Ariyoshi, M, Ohki, I, Tochio, H, Iwai, K, Shirakawa, M. | | 登録日 | 2017-07-31 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Cooperative Domain Formation by Homologous Motifs in HOIL-1L and SHARPIN Plays A Crucial Role in LUBAC Stabilization.
Cell Rep, 23, 2018
|
|
3WDS
 
 | |
