2RO0
 
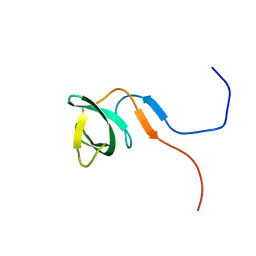 | | Solution structure of the knotted tudor domain of the yeast histone acetyltransferase, Esa1 | | 分子名称: | Histone acetyltransferase ESA1 | | 著者 | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | 登録日 | 2008-03-01 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
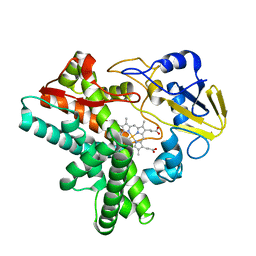
1EHF
 
 | |
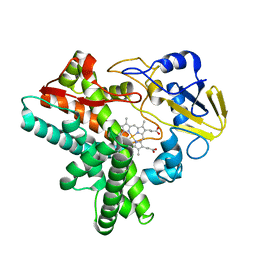
1EHE
 
 | |
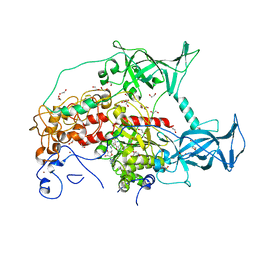
3SWR
 
 | |
1EHG
 
 | |
1MGT
 
 | | CRYSTAL STRUCTURE OF O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM HYPERTHERMOPHILIC ARCHAEON PYROCOCCUS KODAKARAENSIS STRAIN KOD1 | | 分子名称: | PROTEIN (O6-METHYLGUANINE-DNA METHYLTRANSFERASE), SULFATE ION | | 著者 | Hashimoto, H, Inoue, T, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | 登録日 | 1999-01-12 | | 公開日 | 2000-01-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase.
J.Mol.Biol., 292, 1999
|
|
4GEL
 
 | | Crystal structure of Zucchini | | 分子名称: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | 著者 | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.756 Å) | | 主引用文献 | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
5T0U
 
 | | CTCF ZnF2-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
4EW0
 
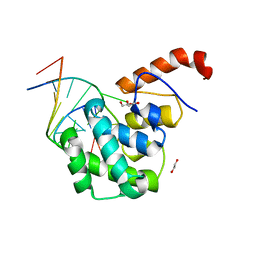 | | mouse MBD4 glycosylase domain in complex with a G:5hmU (5-hydroxymethyluracil) mismatch | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*(5HU)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2012-04-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4GEM
 
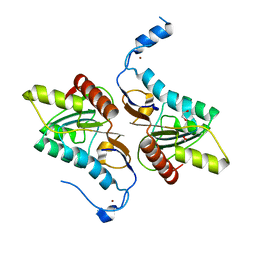 | | Crystal structure of Zucchini (K171A) | | 分子名称: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | 著者 | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4EVV
 
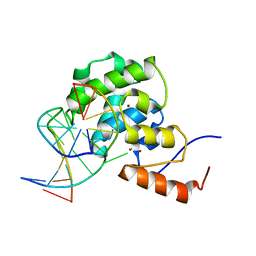 | | mouse MBD4 glycosylase domain in complex with a G:T mismatch | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*TP*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2012-04-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
5T00
 
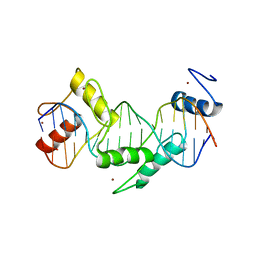 | | Human CTCF ZnF3-7 and methylated DNA complex | | 分子名称: | DNA (5'-GCCAGCAGGGGG(5CM)GCTA-3'), DNA (5'-TAG(5CM)GCCCCCTGCTGGC-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
4EW4
 
 | | mouse MBD4 glycosylase domain in complex with DNA containing a ribose sugar | | 分子名称: | DNA (5'-D(*CP*CP*AP*TP*GP*(3DR)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4, ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2012-04-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.791 Å) | | 主引用文献 | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4FNC
 
 | | Human TDG in a post-reactive complex with 5-hydroxymethyluracil (5hmU) | | 分子名称: | 5-HYDROXYMETHYL URACIL, DNA (28-MER), DNA (29-MER), ... | | 著者 | Hashimoto, H, Hong, S, Bhagwat, A.S, Zhang, X, Cheng, X. | | 登録日 | 2012-06-19 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4GEN
 
 | | Crystal structure of Zucchini (monomer) | | 分子名称: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | 著者 | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
6DNZ
 
 | | Trypanosoma brucei PRMT1 enzyme-prozyme heterotetrameric complex with AdoHcy | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine N-methyltransferase, putative, ... | | 著者 | Hashimoto, H, Kafkova, L, Jordan, K, Read, L.K, Debler, E.W. | | 登録日 | 2018-06-08 | | 公開日 | 2019-06-12 | | 最終更新日 | 2020-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.384 Å) | | 主引用文献 | Structural Basis of Protein Arginine Methyltransferase Activation by a Catalytically Dead Homolog (Prozyme).
J.Mol.Biol., 432, 2020
|
|
5KE6
 
 | |
5KEB
 
 | |
5K5I
 
 | |
5K5L
 
 | |
5K5H
 
 | |
5KE8
 
 | |
5K5J
 
 | |
5KE9
 
 | |
5KEA
 
 | |
