4TNA
 
 | |
2LD1
 
 | |
6R1T
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | 分子名称: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.02 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R25
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.61 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
3NF9
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 1,2-ETHANEDIOL, 5-[(6-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-10 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NFA
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 6-[(5-bromo-2,3-dioxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-10 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF6
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 1,2-ETHANEDIOL, 5-[(2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF7
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 1,2-ETHANEDIOL, 5-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF8
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 6-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
2LBM
 
 | |
2JM1
 
 | |
7V96
 
 | | Telomeric Dinucleosome | | 分子名称: | DNA (275-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Soman, A. | | 登録日 | 2021-08-24 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9K
 
 | | Telomeric tetranucleosome | | 分子名称: | DNA (539-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Soman, A. | | 登録日 | 2021-08-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (8.1 Å) | | 主引用文献 | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9J
 
 | | Telomeric trinucleosome | | 分子名称: | DNA (408-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Soman, A. | | 登録日 | 2021-08-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7VA4
 
 | | Telomeric tetranucleosome in open state | | 分子名称: | DNA (539-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Soman, A. | | 登録日 | 2021-08-27 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (14 Å) | | 主引用文献 | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V90
 
 | | Telomeric mononucleosome | | 分子名称: | DNA (145-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Soman, A. | | 登録日 | 2021-08-24 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9C
 
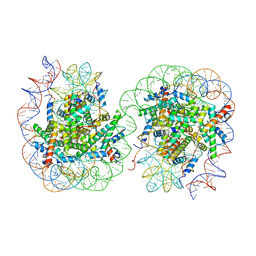 | | Telomeric Dinucleosome in open state | | 分子名称: | DNA (275-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Soman, A. | | 登録日 | 2021-08-24 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9S
 
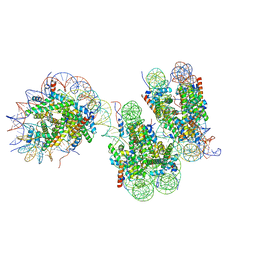 | | Telomeric trinucleosome in open state | | 分子名称: | DNA (408-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Soman, A. | | 登録日 | 2021-08-26 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
1MEY
 
 | |
