6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | 著者 | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | 登録日 | 2020-04-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-22 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3LQQ
 
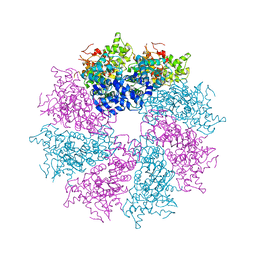 | | Structure of the CED-4 Apoptosome | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Qi, S, Pang, Y, Shi, Y, Yan, N, Liu, Q. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.534 Å) | | 主引用文献 | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
3LQR
 
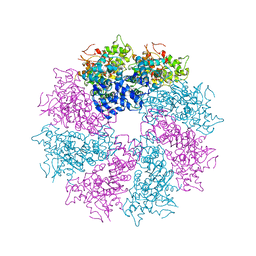 | | Structure of CED-4:CED-3 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Qi, S, Pang, Y, Shi, Y, Yan, N. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.896 Å) | | 主引用文献 | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
2BPR
 
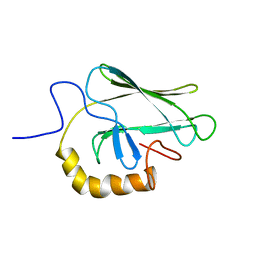 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, 25 STRUCTURES | | 分子名称: | DNAK | | 著者 | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | 登録日 | 1998-08-11 | | 公開日 | 1999-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | 著者 | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | 登録日 | 2000-03-17 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1QX8
 
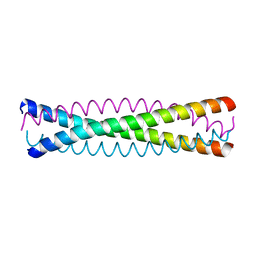 | | Crystal structure of a five-residue deletion mutant of the Rop protein | | 分子名称: | Regulatory protein ROP | | 著者 | Glykos, N.M, Vlassi, M, Papanikolaou, Y, Kotsifaki, D, Cesareni, G, Kokkinidis, M. | | 登録日 | 2003-09-04 | | 公開日 | 2004-09-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Loopless Rop: structure and dynamics of an engineered homotetrameric variant of the repressor of primer protein.
Biochemistry, 45, 2006
|
|
3UDB
 
 | | Crystal structure of SnRK2.6 | | 分子名称: | CHLORIDE ION, Serine/threonine-protein kinase SRK2E | | 著者 | Xie, T, Ren, R, Pang, Y, Yan, C. | | 登録日 | 2011-10-27 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.567 Å) | | 主引用文献 | Molecular mechanism for the inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by the protein phosphatase ABI1
to be published
|
|
1C7S
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT D539A COMPLEXED WITH DI-N-ACETYL-BETA-D-GLUCOSAMINE (CHITOBIASE) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | 著者 | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | 登録日 | 2000-03-14 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1CKR
 
 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | 分子名称: | HEAT SHOCK SUBSTRATE BINDING DOMAIN OF HSC-70 | | 著者 | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | 登録日 | 1999-04-22 | | 公開日 | 1999-04-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
1BPR
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNAK | | 著者 | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | 登録日 | 1998-08-11 | | 公開日 | 1999-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
5ZU4
 
 | |
6IUL
 
 | |
7HSC
 
 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | 分子名称: | PROTEIN (HEAT SHOCK COGNATE 70 KD PROTEIN 1) | | 著者 | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | 登録日 | 1999-05-03 | | 公開日 | 1999-05-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
8GUE
 
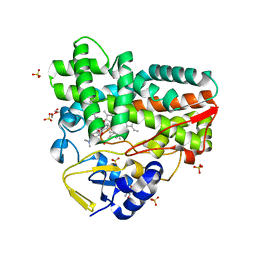 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | 著者 | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | 登録日 | 2022-09-11 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
8DQU
 
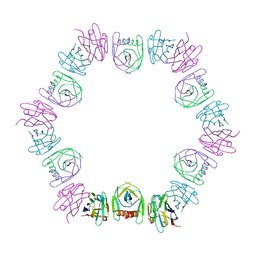 | | Nanobody bound SARS-CoV-2 Nsp9 | | 分子名称: | Nanobody, Non-structural protein 9 | | 著者 | Littler, D.R, Gully, B.S, Rossjohn, J. | | 登録日 | 2022-07-20 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.45003176 Å) | | 主引用文献 | Inside-out: Antibody-binding reveals potential folding hinge-points within the SARS-CoV-2 replication co-factor nsp9.
Plos One, 18, 2023
|
|
3MQ0
 
 | |
5DCC
 
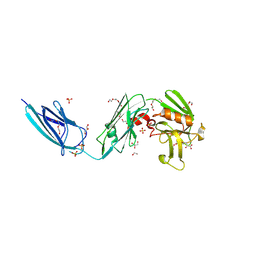 | | X-RAY CRYSTAL STRUCTURE OF a TEBIPENEM ADDUCT OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | 分子名称: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | 登録日 | 2015-08-23 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.451 Å) | | 主引用文献 | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5DC2
 
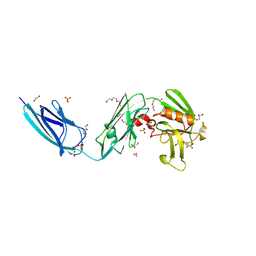 | | X-RAY CRYSTAL STRUCTURE OF A ENZYMATICALLY DEGRADED BIAPENEM-ADDUCT OF L,D-TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | 分子名称: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | 登録日 | 2015-08-23 | | 公開日 | 2016-10-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.182 Å) | | 主引用文献 | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | 分子名称: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | 著者 | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | 登録日 | 2021-02-21 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
6L10
 
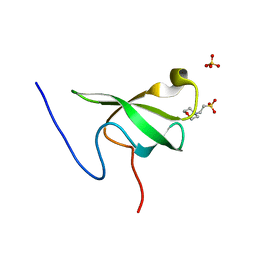 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
8ZC2
 
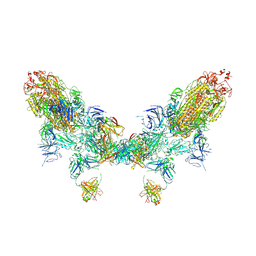 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (7.82 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC6
 
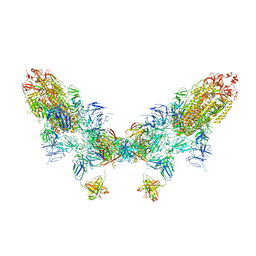 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (6.85 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.69 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
