6Z6G
 
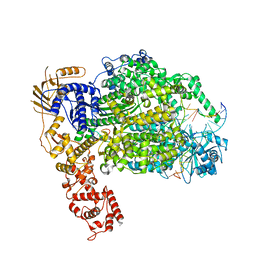 | | Cryo-EM structure of La Crosse virus polymerase at pre-initiation stage | | 分子名称: | 3'vRNA 1-16, 5'vRNA 1-10, 5'vRNA 9-16, ... | | 著者 | Arragain, B, Effantin, G, Gerlach, P, Reguera, J, Schoehn, G, Cusack, S, Malet, H. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
7ZCH
 
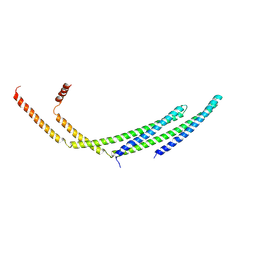 | | CHMP2A-CHMP3 heterodimer (410 Angstrom diameter) | | 分子名称: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | 著者 | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | 登録日 | 2022-03-28 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZCG
 
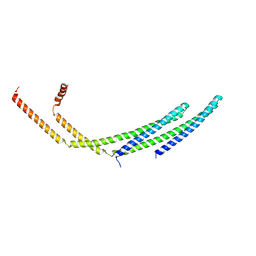 | | CHMP2A-CHMP3 heterodimer (430 Angstrom diameter) | | 分子名称: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | 著者 | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | 登録日 | 2022-03-28 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6YN6
 
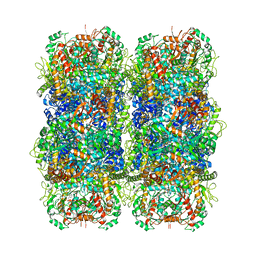 | | Inducible lysine decarboxylase LdcI stacks, pH 5.7 | | 分子名称: | Inducible lysine decarboxylase | | 著者 | Felix, J, Jessop, M, Desfosses, A, Effantin, G, Gutsche, I. | | 登録日 | 2020-04-10 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | 分子名称: | Inducible lysine decarboxylase | | 著者 | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | 登録日 | 2020-04-10 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ZQP
 
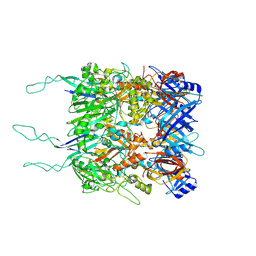 | | Tail tip of siphophage T5 : open cone after interaction with bacterial receptor FhuA | | 分子名称: | Probable baseplate hub protein, Probable tape measure protein | | 著者 | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | 登録日 | 2022-05-02 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZHJ
 
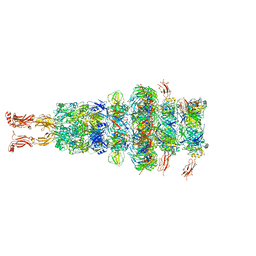 | | Tail tip of siphophage T5 : tip proteins | | 分子名称: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | 著者 | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | 登録日 | 2022-04-06 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZN2
 
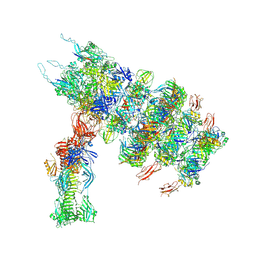 | | Tail tip of siphophage T5 : full complex after interaction with its bacterial receptor FhuA | | 分子名称: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | 著者 | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | 登録日 | 2022-04-20 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZN4
 
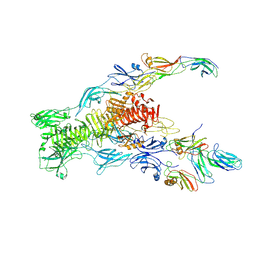 | | Tail tip of siphophage T5 : bent fibre after interaction with its bacterial receptor FhuA | | 分子名称: | Probable baseplate hub protein, Probable central straight fiber | | 著者 | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | 登録日 | 2022-04-20 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.32 Å) | | 主引用文献 | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZQB
 
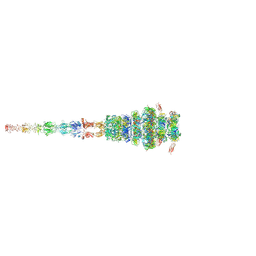 | | Tail tip of siphophage T5 : full structure | | 分子名称: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | 著者 | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | 登録日 | 2022-04-29 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZLV
 
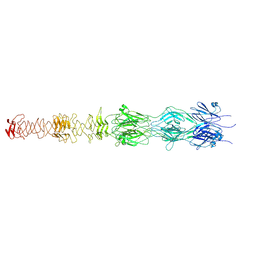 | | Tail tip of siphophage T5 : central fibre protein pb4 | | 分子名称: | Probable central straight fiber | | 著者 | Linares, R, Arnaud, C.A, Effantin, G, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | 登録日 | 2022-04-15 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.22 Å) | | 主引用文献 | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
8B14
 
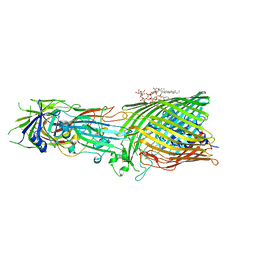 | | T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA | | 分子名称: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, FhuA iron-ferrichrome transporter, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate, ... | | 著者 | Degroux, S, Effantin, G, Linares, R, Schoehn, G, Breyton, C. | | 登録日 | 2022-09-09 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger.
J.Virol., 97, 2023
|
|
8A1R
 
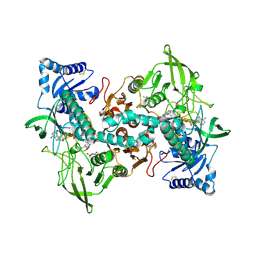 | | cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-[3-[[[(1~{R},2~{R},3~{R},5~{S})-2,6,6-trimethyl-3-bicyclo[3.1.1]heptanyl]amino]methyl]indol-1-yl]oxane-3,4,5-triol, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | 著者 | Ardini, M, Angelucci, F, Fata, F, Gabriele, F, Effantin, G, Ling, W, Williams, D.L, Petukhova, V.Z, Petukhov, P.A. | | 登録日 | 2022-06-01 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Non-covalent inhibitors of thioredoxin glutathione reductase with schistosomicidal activity in vivo.
Nat Commun, 14, 2023
|
|
8BCU
 
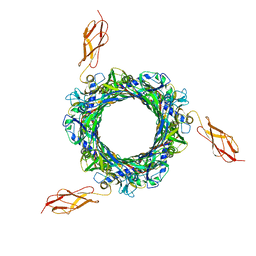 | |
8BCP
 
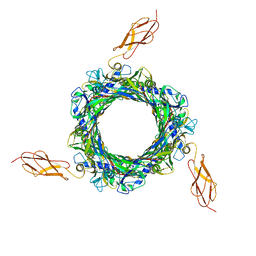 | |
6Y1Z
 
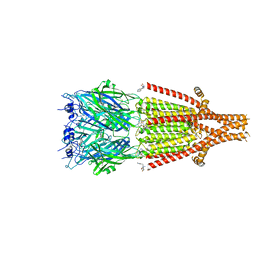 | | Mouse serotonin 5HT3 receptor in complex with palonosetron | | 分子名称: | (3~{a}~{S})-2-[(3~{S})-1-azabicyclo[2.2.2]octan-3-yl]-3~{a},4,5,6-tetrahydro-3~{H}-benzo[de]isoquinolin-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zarkadas, E, Perot, J, Nury, H. | | 登録日 | 2020-02-14 | | 公開日 | 2020-03-04 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (2.82 Å) | | 主引用文献 | The Binding of Palonosetron and Other Antiemetic Drugs to the Serotonin 5-HT3 Receptor.
Structure, 28, 2020
|
|
7O76
 
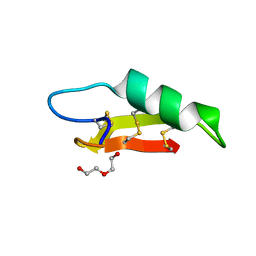 | |
5M1M
 
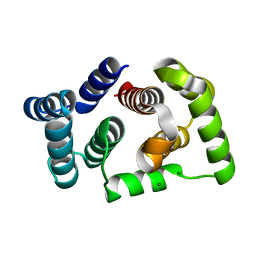 | |
6Z6B
 
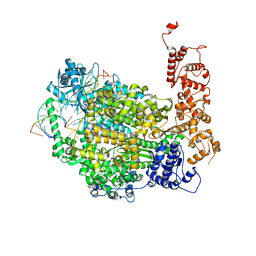 | | Structure of full-length La Crosse virus L protein (polymerase) | | 分子名称: | RNA (5'-R(*GP*CP*UP*AP*CP*UP*AP*A)-3'), RNA (5'-R(P*AP*GP*UP*AP*GP*UP*GP*UP*GP*C)-3'), RNA (5'-R(P*UP*UP*AP*GP*UP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), ... | | 著者 | Cusack, S, Gerlach, P, Reguera, J. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.961 Å) | | 主引用文献 | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
5FL2
 
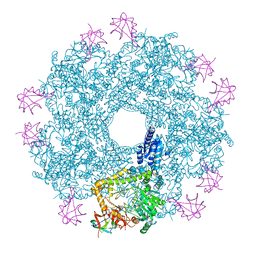 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | 分子名称: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | 著者 | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | 登録日 | 2015-10-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
