5K5I
 
 | |
5KE6
 
 | |
5KEB
 
 | |
3F8I
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group P21 | | 分子名称: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | 著者 | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2008-11-12 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3FDE
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | 著者 | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2008-11-25 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3F8J
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | 分子名称: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | 著者 | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2008-11-12 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
1F0O
 
 | |
1EYU
 
 | |
1YFJ
 
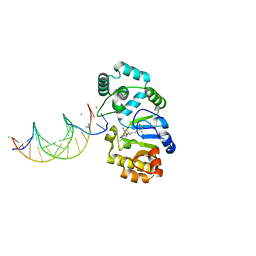 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | 分子名称: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-02 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YF3
 
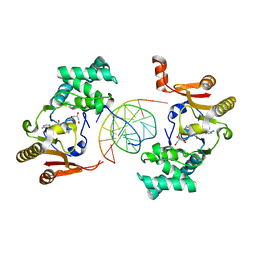 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | 分子名称: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2004-12-30 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YFL
 
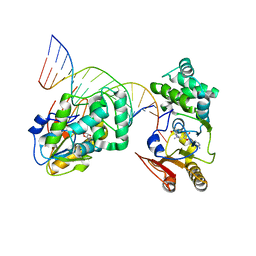 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | 分子名称: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-03 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
2HMY
 
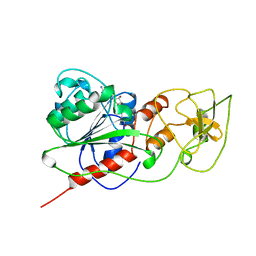 | | BINARY COMPLEX OF HHAI METHYLTRANSFERASE WITH ADOMET FORMED IN THE PRESENCE OF A SHORT NONPSECIFIC DNA OLIGONUCLEOTIDE | | 分子名称: | PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), S-ADENOSYLMETHIONINE | | 著者 | O'Gara, M, Zhang, X, Roberts, R.J, Cheng, X. | | 登録日 | 1999-02-08 | | 公開日 | 1999-03-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure of a binary complex of HhaI methyltransferase with S-adenosyl-L-methionine formed in the presence of a short non-specific DNA oligonucleotide.
J.Mol.Biol., 287, 1999
|
|
2PVI
 
 | |
9MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | 分子名称: | 5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | 著者 | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | 登録日 | 1998-08-07 | | 公開日 | 1998-12-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
6OX2
 
 | | SETD3in Complex with an Actin Peptide with the Target Histidine Fully Methylated | | 分子名称: | 1,2-ETHANEDIOL, Actin Peptide, GLYCEROL, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.089 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
2AOT
 
 | | Histamine Methyltransferase Complexed with the Antihistamine Drug Diphenhydramine | | 分子名称: | Histamine N-methyltransferase, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | 登録日 | 2005-08-14 | | 公開日 | 2005-09-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
6OX4
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.294 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX3
 
 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Lysine | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.785 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX1
 
 | | SETD3 in Complex with an Actin Peptide with Target Histidine Partially Methylated | | 分子名称: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX5
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | 分子名称: | 1,2-ETHANEDIOL, Actin Peptide, Actin-histidine N-methyltransferase, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
5T00
 
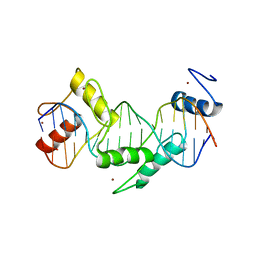 | | Human CTCF ZnF3-7 and methylated DNA complex | | 分子名称: | DNA (5'-GCCAGCAGGGGG(5CM)GCTA-3'), DNA (5'-TAG(5CM)GCCCCCTGCTGGC-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5T01
 
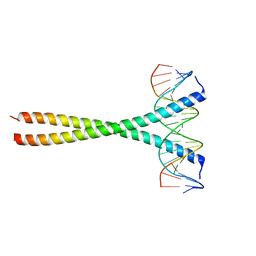 | | Human c-Jun DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*GP*AP*(5CM)P*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*GP*TP*CP*CP*AP*T)-3'), Transcription factor AP-1 | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
6OX0
 
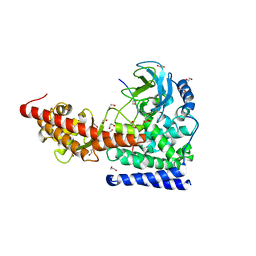 | | SETD3 in Complex with an Actin Peptide with Sinefungin Replacing SAH as Cofactor | | 分子名称: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.755 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
5SZX
 
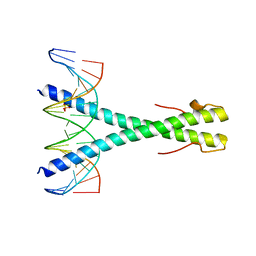 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5T0U
 
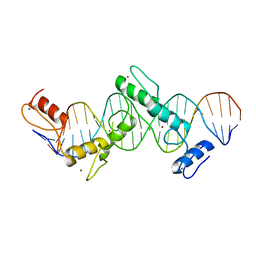 | | CTCF ZnF2-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
