7YUH
 
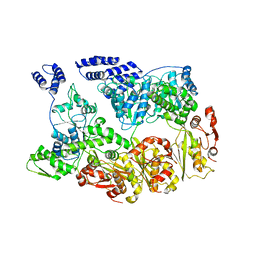 | | MtaLon-Apo for the spiral oligomers of trimer | | 分子名称: | Lon protease | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-17 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUP
 
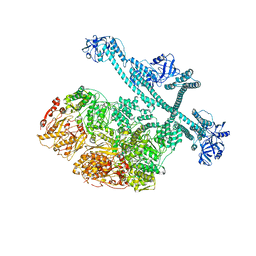 | | MtaLon-Apo for the spiral oligomers of pentamer | | 分子名称: | Lon protease | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-17 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUT
 
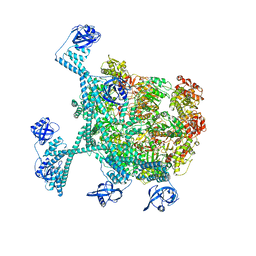 | | MtaLon-Apo for the spiral oligomers of hexamer | | 分子名称: | Lon protease | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-17 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUX
 
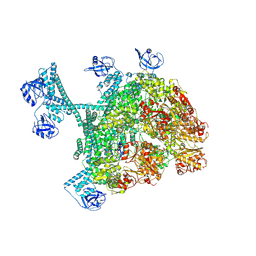 | | MtaLon-ADP for the spiral oligomers of hexamer | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-18 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7FID
 
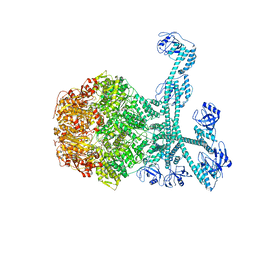 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIZ
 
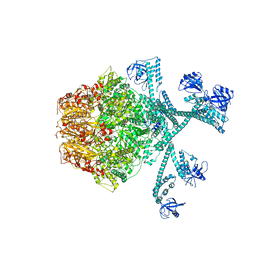 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-08-01 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIE
 
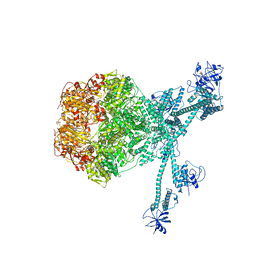 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
2M5V
 
 | |
5WQL
 
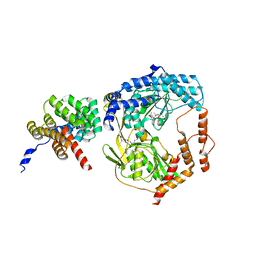 | |
4OM4
 
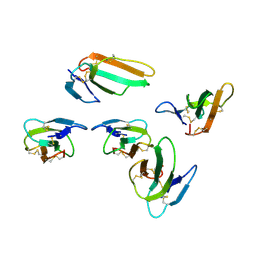 | |
4OM5
 
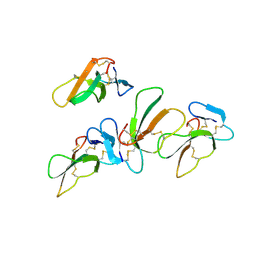 | |
1LEW
 
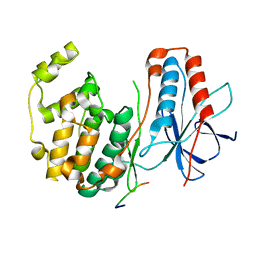 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS NUCLEAR SUBSTRATE MEF2A | | 分子名称: | Mitogen-activated protein kinase 14, Myocyte-specific enhancer factor 2A | | 著者 | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | 登録日 | 2002-04-10 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
1LEZ
 
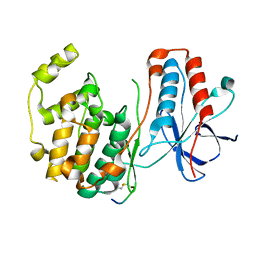 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS ACTIVATOR MKK3B | | 分子名称: | MAP kinase kinase 3b, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | 著者 | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | 登録日 | 2002-04-10 | | 公開日 | 2002-07-10 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
8IKR
 
 | |
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | 著者 | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | 登録日 | 2020-04-28 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
7FD4
 
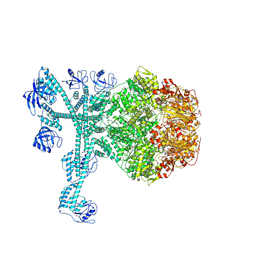 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FD5
 
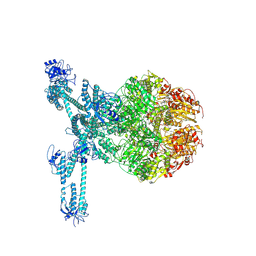 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7CRA
 
 | |
7CR9
 
 | |
4R8F
 
 | |
2PN5
 
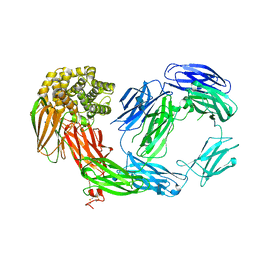 | | Crystal Structure of TEP1r | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | 著者 | Baxter, R.H.G. | | 登録日 | 2007-04-23 | | 公開日 | 2007-07-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.698 Å) | | 主引用文献 | Structural basis for conserved complement factor-like function in the antimalarial protein TEP1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
