8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | 分子名称: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | 登録日 | 2023-12-04 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
7NAC
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAD
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb4 local refinement model | | 分子名称: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L17-A, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAF
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb1-MTD local model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R7O
 
 | |
7R6K
 
 | | State E2 nucleolar 60S ribosomal intermediate - Model for Noc2/Noc3 region | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-22 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R7C
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - L1 stalk local model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-24 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R72
 
 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Spb4 local model | | 分子名称: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-24 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R7A
 
 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Composite model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-24 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R6Q
 
 | | State E2 nucleolar 60S ribosome biogenesis intermediate - Foot region model | | 分子名称: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-23 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4CE4
 
 | | 39S large subunit of the porcine mitochondrial ribosome | | 分子名称: | 16S Ribosomal RNA, ICT1, MRPL13, ... | | 著者 | Greber, B.J, Boehringer, D, Leitner, A, Bieri, P, Voigts-Hoffmann, F, Erzberger, J.P, Leibundgut, M, Aebersold, R, Ban, N. | | 登録日 | 2013-11-08 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Architecture of the Large Subunit of the Mammalian Mitochondrial Ribosome.
Nature, 505, 2014
|
|
6B1E
 
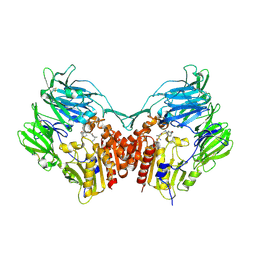 | | The structure of DPP4 in complex with Vildagliptin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(1r,3s,5R,7S)-3-hydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]amino}-1-{(2S)-2-[(E)-iminomethyl]pyrrolidin-1-yl}ethan-1-o ne, ... | | 著者 | Scapin, G. | | 登録日 | 2017-09-18 | | 公開日 | 2017-09-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
6B1O
 
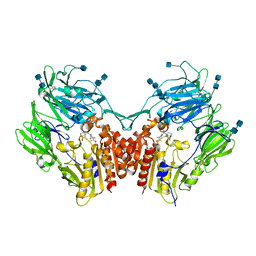 | | The structure of DPP4 in complex with Vildagliptin Analog | | 分子名称: | (2S)-2-amino-1-[(1S,3S,5S)-3-(aminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(1r,3R,5S,7S)-3,5-dihydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G. | | 登録日 | 2017-09-18 | | 公開日 | 2017-09-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
3TY0
 
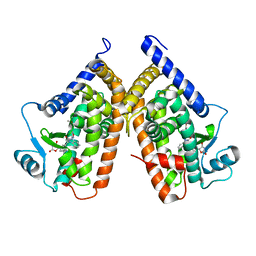 | | Structure of PPARgamma ligand binding domain in complex with (R)-5-(3-((3-(6-methoxybenzo[d]isoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)methyl)phenyl)-5-methyloxazolidine-2,4-dione | | 分子名称: | (5R)-5-(3-{[3-(6-methoxy-1,2-benzoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl]methyl}phenyl)-5-methyl-1,3-oxazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | 著者 | Soisson, S.M, Meinke, P.M, McKeever, B, Liu, W. | | 登録日 | 2011-09-23 | | 公開日 | 2011-11-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Benzimidazolones: a new class of selective peroxisome proliferator-activated receptor gamma (PPAR-gamma) modulators.
J.Med.Chem., 54, 2011
|
|
2P4Y
 
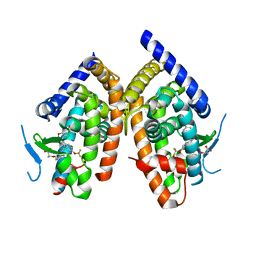 | | Crystal structure of human PPAR-gamma-ligand binding domain complexed with an indole-based modulator | | 分子名称: | (2R)-2-(4-CHLORO-3-{[3-(6-METHOXY-1,2-BENZISOXAZOL-3-YL)-2-METHYL-6-(TRIFLUOROMETHOXY)-1H-INDOL-1-YL]METHYL}PHENOXY)PROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peroxisome proliferator-activated receptor gamma | | 著者 | McKeever, B.M. | | 登録日 | 2007-03-13 | | 公開日 | 2008-01-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The differential interactions of peroxisome proliferator-activated receptor gamma ligands with Tyr473 is a physical basis for their unique biological activities.
Mol.Pharmacol., 73, 2008
|
|
1HD7
 
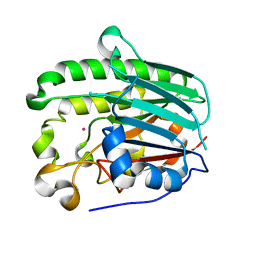 | | A Second Divalent Metal Ion in the Active Site of a New Crystal Form of Human Apurinic/Apyridinimic Endonuclease, Ape1, and its Implications for the Catalytic Mechanism | | 分子名称: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | 著者 | Beernink, P.T, Segelke, B.W, Rupp, B. | | 登録日 | 2000-11-09 | | 公開日 | 2001-02-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
5H7U
 
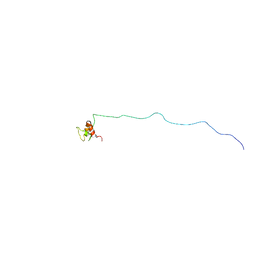 | | NMR structure of eIF3 36-163 | | 分子名称: | Eukaryotic translation initiation factor 3 subunit C | | 著者 | Nagata, T, Obayashi, E. | | 登録日 | 2016-11-21 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
2RVH
 
 | | NMR structure of eIF1 | | 分子名称: | Eukaryotic translation initiation factor eIF-1 | | 著者 | Nagata, T, Obayashi, E, Asano, K. | | 登録日 | 2015-10-16 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5
Cell Rep, 18, 2017
|
|
1E9N
 
 | | A second divalent metal ion in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1, and its implications for the catalytic mechanism | | 分子名称: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | 著者 | Beernink, P.T, Segelke, B.W, Rupp, B. | | 登録日 | 2000-10-24 | | 公開日 | 2001-02-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
