4JAW
 
 | | Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline | | 分子名称: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Lacto-N-biosidase, SULFATE ION, ... | | 著者 | Ito, T, Katayama, T, Stubbs, K.A, Fushinobu, S. | | 登録日 | 2013-02-19 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
5YY9
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | 著者 | Kori, S, Defossez, P.A, Arita, K. | | 登録日 | 2017-12-08 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5YYA
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | 著者 | Kori, S, Defossez, P.A, Arita, K. | | 登録日 | 2017-12-08 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5ZE9
 
 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | 登録日 | 2018-02-27 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5ZEA
 
 | | Crystal structure of the nucleotide-free mutant A3B3 | | 分子名称: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | 著者 | Maruyama, S, Suzuki, K, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | 登録日 | 2018-02-27 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.384 Å) | | 主引用文献 | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5Z5E
 
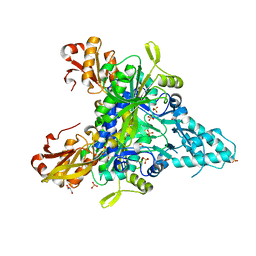 | |
6AI2
 
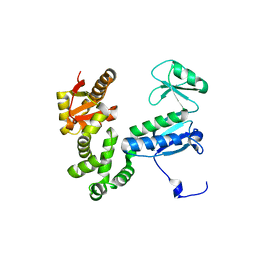 | |
6AI1
 
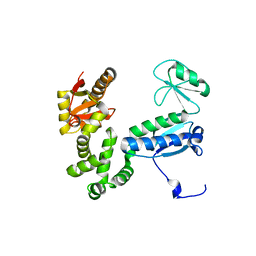 | |
2DF3
 
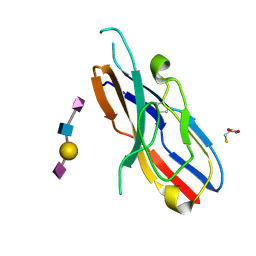 | |
8WU5
 
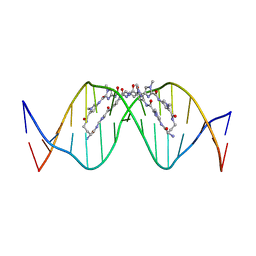 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | 分子名称: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | 著者 | Abe, K, Takeda, K, Sugiyama, H. | | 登録日 | 2023-10-20 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
4EOX
 
 | |
6OW7
 
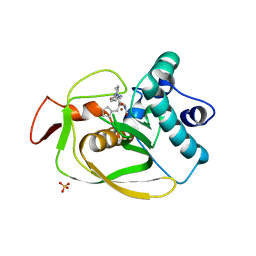 | | X-ray Structure of Polypeptide Deformylase with a Piperazic Acid | | 分子名称: | (3S)-2-{(2R)-2-(cyclopentylmethyl)-3-[formyl(hydroxy)amino]propanoyl}-N-(pyridin-2-yl)hexahydropyridazine-3-carboxamide, NICKEL (II) ION, Peptide deformylase, ... | | 著者 | Campobasso, N, Spletstoser, J, Ward, P. | | 登録日 | 2019-05-09 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OW2
 
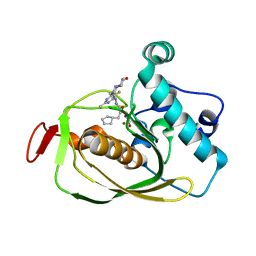 | | X-ray Structure of Polypeptide Deformylase | | 分子名称: | (2R)-2-(cyclopentylmethyl)-N'-{5-fluoro-6-[(9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]-2-methylpyrimidin-4-yl}-3-[hydroxy(hydroxymethyl)amino]propanehydrazide, NICKEL (II) ION, Peptide deformylase | | 著者 | Campobasso, N, Spletstoser, J, Ward, P. | | 登録日 | 2019-05-09 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3T8X
 
 | | Crystal structure of human CD1b in complex with synthetic antigenic diacylsulfoglycolipid SGL12 and endogenous spacer | | 分子名称: | 2-O-sulfo-alpha-D-glucopyranosyl 2-O-hexadecanoyl-3-O-[(2E,4S,6S,8S)-2,4,6,8-tetramethyltetracos-2-enoyl]-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Garcia-Alles, L.F, Maveyraud, L, Mourey, L, Julien, S. | | 登録日 | 2011-08-02 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural reorganization of the human CD1b Antigen-binding groove for presentation of mycobacterial sulfoglycolipids
To be Published
|
|
