1WJA
 
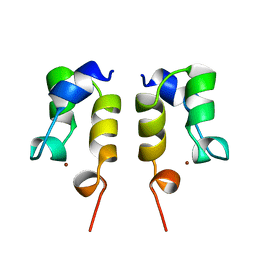 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
7SD4
 
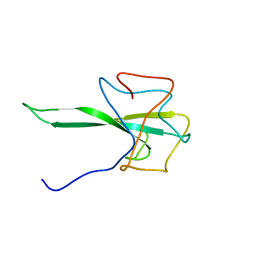 | | SARS-CoV-2 Nucleocapsid N-terminal domain (N-NTD) protein | | 分子名称: | Nucleoprotein | | 著者 | Sarkar, S, Runge, B, Russell, R.W, Calero, D, Zeinalilathori, S, Quinn, C.M, Lu, M, Calero, G, Gronenborn, A.M, Polenova, T. | | 登録日 | 2021-09-29 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Atomic-Resolution Structure of SARS-CoV-2 Nucleocapsid Protein N-Terminal Domain.
J.Am.Chem.Soc., 144, 2022
|
|
1N02
 
 | |
6GAT
 
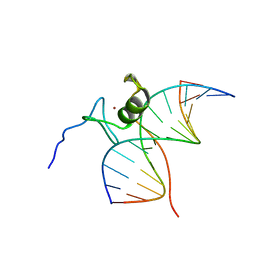 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, REGULARIZED MEAN STRUCTURE | | 分子名称: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | 著者 | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | 登録日 | 1997-11-07 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
7RIK
 
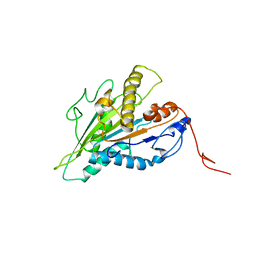 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | 分子名称: | Kinesin-1 heavy chain | | 著者 | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | 登録日 | 2021-07-20 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
7R7Q
 
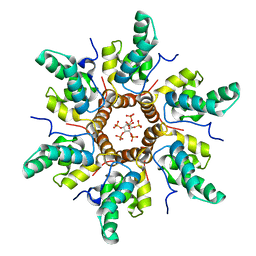 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | 分子名称: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | 著者 | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | 登録日 | 2021-06-25 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | 分子名称: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | 著者 | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | 登録日 | 2021-06-25 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
5HIR
 
 | |
1SAK
 
 | |
1SAE
 
 | |
1SAF
 
 | |
1SAL
 
 | |
4Z8L
 
 | | Crystal structure of DCAF1/SIV-MND VPX/MND SAMHD1 NTD ternary complex | | 分子名称: | Protein VPRBP, SAM domain and HD domain-containing protein, Vpx protein, ... | | 著者 | Koharudin, L.M, Wu, Y, Calero, G, Ahn, J, Gronenborn, A.M. | | 登録日 | 2015-04-09 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile alpha Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors.
J.Biol.Chem., 290, 2015
|
|
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | 分子名称: | HIV-1 capsid protein | | 著者 | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | 登録日 | 2020-05-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6XQJ
 
 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | 分子名称: | Protein Vpr,UV excision repair protein RAD23 homolog A, ZINC ION | | 著者 | Byeon, I.-J.L, Calero, G, Wu, Y, Byeon, C.H, Gronenborn, A.M. | | 登録日 | 2020-07-09 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
3EZE
 
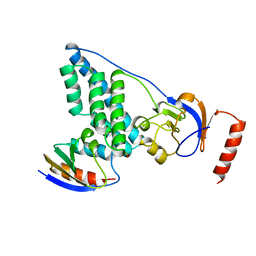 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHITE ION, PROTEIN (PHOSPHOTRANSFERASE SYSTEM, ENZYME I), ... | | 著者 | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1998-11-04 | | 公開日 | 1998-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
3EZA
 
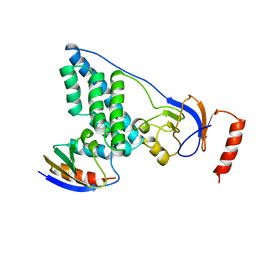 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR, PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1998-11-03 | | 公開日 | 1999-05-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
3EZB
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI | | 分子名称: | PROTEIN (PHOSPHOCARRIER PROTEIN HPR), PROTEIN (PHOSPHOTRANSFER SYSTEM, ENZYME I) | | 著者 | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1998-11-03 | | 公開日 | 1999-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
5K79
 
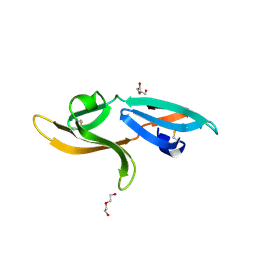 | | Structure and anti-HIV activity of CYT-CVNH, a new cyanovirin-n homolog | | 分子名称: | 1,2-ETHANEDIOL, Cyanovirin-N domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Matei, E, Basu, R, Furey, W, Shi, J, Calnan, C, Aiken, C, Gronenborn, A.M. | | 登録日 | 2016-05-25 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and Glycan Binding of a New Cyanovirin-N Homolog.
J.Biol.Chem., 291, 2016
|
|
3S5V
 
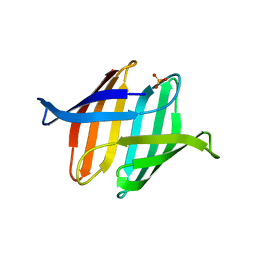 | |
3S60
 
 | |
3S5X
 
 | |
1HUN
 
 | |
3LHC
 
 | | Crystal structure of cyanovirin-n swapping domain b mutant | | 分子名称: | Cyanovirin-N, PHOSPHATE ION, SODIUM ION | | 著者 | Matei, E, Zheng, A, Furey, W, Rose, J, Aiken, C, Gronenborn, A.M. | | 登録日 | 2010-01-21 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization.
J.Biol.Chem., 285, 2010
|
|
3OBL
 
 | |
