3TJ4
 
 | | Crystal structure of an enolase from agrobacterium tumefaciens (efi target efi-502087) no mg | | 分子名称: | CHLORIDE ION, GLYCEROL, Mandelate racemase, ... | | 著者 | Vetting, M.W, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-08-23 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of an enolase from agrobacterium tumefaciens (efi target efi-502087) no mg
to be published
|
|
3TK4
 
 | |
3RR1
 
 | | Crystal structure of enolase PRK14017 (target EFI-500653) from Ralstonia pickettii 12J | | 分子名称: | CHLORIDE ION, D-MALATE, Putative D-galactonate dehydratase | | 著者 | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-04-28 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of enolase PRK14017 from Ralstonia pickettii
To be Published
|
|
3RGU
 
 | | Structure of Fap-NRa at pH 5.0 | | 分子名称: | Fimbriae-associated protein Fap1, alpha-D-glucopyranose | | 著者 | Garnett, J.A, Matthews, S.J. | | 登録日 | 2011-04-09 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insight into the role of Streptococcus parasanguinis Fap1 within oral biofilm formation.
Biochem.Biophys.Res.Commun., 417, 2012
|
|
3RHG
 
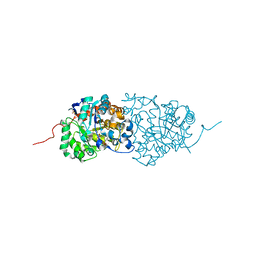 | | Crystal structure of amidohydrolase pmi1525 (target efi-500319) from proteus mirabilis hi4320 | | 分子名称: | BENZOIC ACID, CACODYLATE ION, Putative phophotriesterase, ... | | 著者 | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-04-11 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
3R10
 
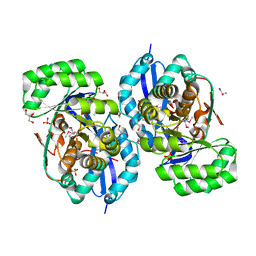 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Mg complex | | 分子名称: | Enzyme of enolase superfamily, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-03-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RO6
 
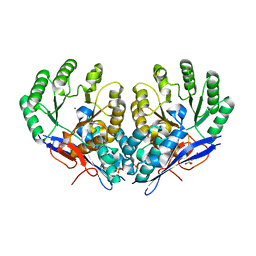 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg ion | | 分子名称: | GLYCEROL, MAGNESIUM ION, Putative chloromuconate cycloisomerase, ... | | 著者 | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | 登録日 | 2011-04-25 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3THU
 
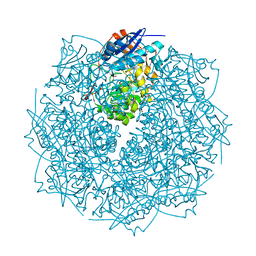 | | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-08-19 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg
to be published
|
|
3TJI
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ENTEROBACTER sp. 638 (EFI TARGET EFI-501662) with BOUND MG | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-08-24 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of an enolase from enterobacter sp. 638 (efi target efi-501662) with boung mg
to be published
|
|
3TWB
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and gluconic acid | | 分子名称: | CHLORIDE ION, D-gluconic acid, GLYCEROL, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-09-21 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3TOY
 
 | | CRYSTAL STRUCTURE OF ENOLASE BRADO_4202 (TARGET EFI-501651) FROM Bradyrhizobium sp. ORS278 WITH CALCIUM AND ACETATE BOUND | | 分子名称: | ACETATE ION, CALCIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | 著者 | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-09-06 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF MANDELATE RACEMASE FROM Bradyrhizobium sp. ORS278
To be Published
|
|
3U11
 
 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | 登録日 | 2011-09-29 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
3TWA
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and glycerol | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-09-21 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3PSG
 
 | |
3Q72
 
 | | Crystal Structure of Rad G-domain-GTP Analog Complex | | 分子名称: | CALCIUM ION, GTP-binding protein RAD, MAGNESIUM ION, ... | | 著者 | Sasson, Y, Navon-Perry, L, Hirsch, J.A. | | 登録日 | 2011-01-04 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.655 Å) | | 主引用文献 | RGK Family G-Domain:GTP Analog Complex Structures and Nucleotide-Binding Properties.
J.Mol.Biol., 413, 2011
|
|
3Q7P
 
 | |
3QIL
 
 | |
3QKE
 
 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter Salexigens complexed with Mg and D-Gluconate | | 分子名称: | D-gluconic acid, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | 著者 | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-02-01 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Cystal structure of D-mannonate dehydratase from Chromohalobacter salexigens complexed with Mg and D-Gluconate
To be Published
|
|
3QNM
 
 | | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function | | 分子名称: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | 著者 | Matthew, M.W, Ramagopal, U.A, Toro, R, Dickey, M, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2011-02-08 | | 公開日 | 2011-03-30 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function
To be Published
|
|
3TNI
 
 | |
3TWE
 
 | | Crystal Structure of the de novo designed peptide alpha4H | | 分子名称: | ACETYL GROUP, TRIETHYLENE GLYCOL, alpha4H | | 著者 | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | 登録日 | 2011-09-21 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TWG
 
 | |
3TVW
 
 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 4 | | 分子名称: | Acetyl-CoA carboxylase, [4-(2H-chromen-3-ylmethyl)piperazin-1-yl]-[3-(1H-pyrazol-5-yl)phenyl]methanone | | 著者 | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | 登録日 | 2011-09-20 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
3TNH
 
 | | CDK9/cyclin T in complex with CAN508 | | 分子名称: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-T1, Cyclin-dependent kinase 9 | | 著者 | Baumli, S, Hole, A.J, Endicott, J.A. | | 登録日 | 2011-09-01 | | 公開日 | 2012-02-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.202 Å) | | 主引用文献 | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
3TW9
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative dehydratase | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-09-21 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
