7KNI
 
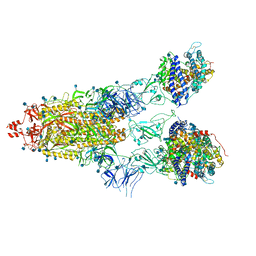 | | Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNE
 
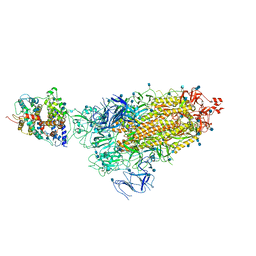 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNB
 
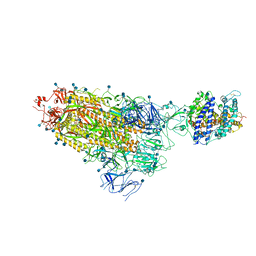 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMS
 
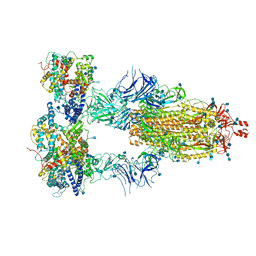 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMZ
 
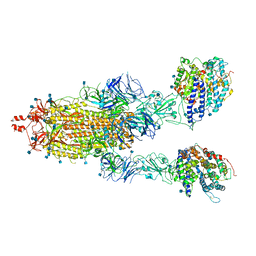 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KOV
 
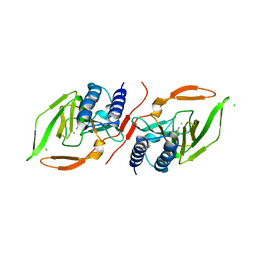 | |
7LRT
 
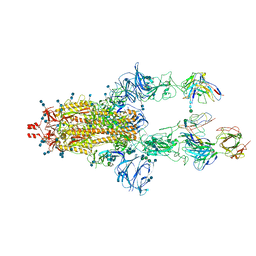 | |
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2021-02-13 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
7LRS
 
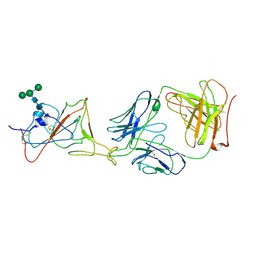 | |
7KMB
 
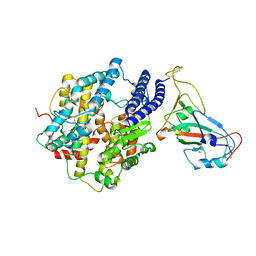 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-02 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNH
 
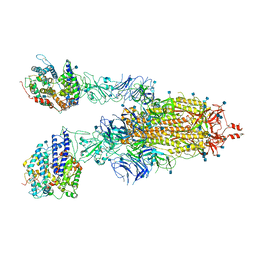 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KJP
 
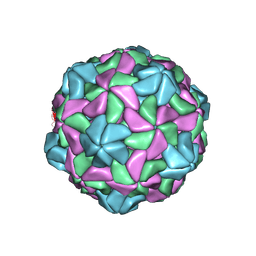 | |
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | 著者 | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | 登録日 | 2021-04-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.427 Å) | | 主引用文献 | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
7MM0
 
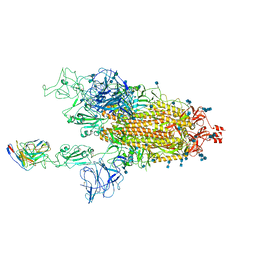 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | 著者 | Zhou, T, Tsybovsky, T, Kwong, P.D. | | 登録日 | 2021-04-29 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7MLZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | 分子名称: | B1-182.1 Fab heavy chain, B1-182.1 Fab light chain, Spike protein S1, ... | | 著者 | Zhou, T, Tsybovsky, T, Kwong, P.D. | | 登録日 | 2021-04-29 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
5WUX
 
 | | TNFalpha-certolizumab Fab | | 分子名称: | Tumor necrosis factor alpha, heavy, light | | 著者 | Heo, Y.S, Lee, J.U. | | 登録日 | 2016-12-21 | | 公開日 | 2017-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
7S0D
 
 | |
7S0E
 
 | |
7S0B
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | 著者 | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2021-08-30 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
7S0C
 
 | |
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BUC
 
 | | Crystal structure of EHMT2 SET domain in complex with compound 13 | | 分子名称: | Histone-lysine N-methyltransferase EHMT2, N2-[4-methoxy-3-(2,3,4,7-tetrahydro-1H-azepin-5-yl)phenyl]-N4,6-dimethyl-pyrimidine-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | 著者 | Suzuki, M, Mizuno, M, Katayama, K. | | 登録日 | 2020-04-06 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of novel histone lysine methyltransferase G9a/GLP (EHMT2/1) inhibitors: Design, synthesis, and structure-activity relationships of 2,4-diamino-6-methylpyrimidines.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7BTV
 
 | | Crystal structure of EHMT2 SET domain in complex with compound 5. | | 分子名称: | Histone-lysine N-methyltransferase EHMT2, N~2~-{4-methoxy-3-[3-(pyrrolidin-1-yl)propoxy]phenyl}-N~4~,6-dimethylpyrimidine-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | 著者 | Suzuki, M, Mizuno, T, Katayama, K. | | 登録日 | 2020-04-03 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of novel histone lysine methyltransferase G9a/GLP (EHMT2/1) inhibitors: Design, synthesis, and structure-activity relationships of 2,4-diamino-6-methylpyrimidines.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5B6O
 
 | | Crystal structure of MS8104 | | 分子名称: | 3C-like proteinase | | 著者 | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2016-05-31 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8ZJF
 
 | | Cryo-EM structure of human integrin alpha-E beta-7 | | 分子名称: | CALCIUM ION, Integrin alpha-E, Integrin beta-7, ... | | 著者 | Akasaka, H, Nureki, O, Kise, Y. | | 登録日 | 2024-05-14 | | 公開日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structure of I domain-containing integrin alpha E beta 7.
Biochem.Biophys.Res.Commun., 721, 2024
|
|
