5URM
 
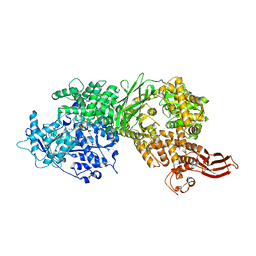 | | Crystal structure of human BRR2 in complex with T-1206548 | | 分子名称: | 3-(5-{[(2R)-5-amino-2-cyclohexyl-7-oxo-2,3-dihydro-7H-[1,3,4]thiadiazolo[3,2-a]pyrimidin-6-yl]methyl}furan-2-yl)benzoic acid, U5 small nuclear ribonucleoprotein 200 kDa helicase | | 著者 | Klein, M.G, Tjhen, R, Qin, L. | | 登録日 | 2017-02-11 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URJ
 
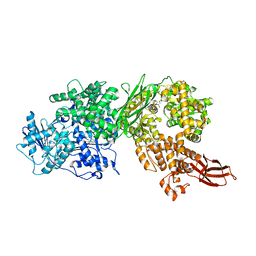 | | Crystal structure of human BRR2 in complex with T-3905516 | | 分子名称: | 6-benzyl-3-[(2R)-2-(3-fluoropyridin-2-yl)-6-methyl-3,4-dihydro-2H-1-benzopyran-7-yl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(3H,8H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | 著者 | Klein, M.G, Tjhen, R, Qin, L. | | 登録日 | 2017-02-10 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URK
 
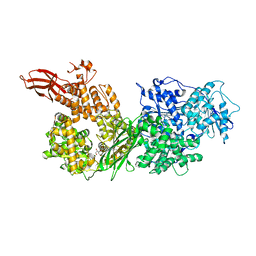 | | Crystal structure of human BRR2 in complex with T-3935799 | | 分子名称: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | 著者 | Qin, L, Tjhen, R, Klein, M.G. | | 登録日 | 2017-02-10 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
6A3I
 
 | | Levoglucosan dehydrogenase, complex with NADH and levoglucosan | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Levoglucosan, Putative dehydrogenase | | 著者 | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | 登録日 | 2018-06-15 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6A3F
 
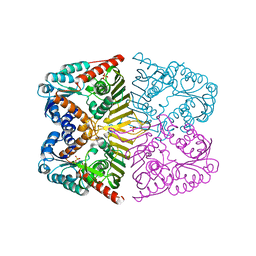 | | Levoglucosan dehydrogenase, apo form | | 分子名称: | Putative dehydrogenase, SULFATE ION | | 著者 | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | 登録日 | 2018-06-15 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6A3G
 
 | | Levoglucosan dehydrogenase, complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative dehydrogenase | | 著者 | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | 登録日 | 2018-06-15 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
1RDB
 
 | |
5Y5F
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y0C
 
 | | Crystal Structure of the human nucleosome at 2.09 angstrom resolution | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | 登録日 | 2017-07-16 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.087 Å) | | 主引用文献 | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
1RDC
 
 | |
8H3M
 
 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | 著者 | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | 登録日 | 2022-10-09 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | 著者 | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | 登録日 | 2022-10-09 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
5Y0D
 
 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | 登録日 | 2017-07-16 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5Y5G
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5GQG
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex | | 分子名称: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | 著者 | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | 登録日 | 2016-08-07 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | 著者 | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | 登録日 | 2017-04-10 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
5GQF
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex | | 分子名称: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | 登録日 | 2016-08-07 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
7CJI
 
 | | Photosystem II structure in the S1 state | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Li, H, Shen, J.-R, Suga, M. | | 登録日 | 2020-07-11 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7CJJ
 
 | | Photosystem II structure in the S2 state | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Li, H, Shen, J.-R, Suga, M. | | 登録日 | 2020-07-11 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7COU
 
 | | Structure of cyanobacterial photosystem II in the dark S1 state | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Li, H, Shen, J.-R, Suga, M. | | 登録日 | 2020-08-05 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
1ENK
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | 分子名称: | ENDONUCLEASE V | | 著者 | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | 登録日 | 1994-08-08 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
5GQC
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form | | 分子名称: | CALCIUM ION, Lacto-N-biosidase, SODIUM ION | | 著者 | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | 登録日 | 2016-08-07 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
1ENI
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | 分子名称: | ENDONUCLEASE V | | 著者 | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | 登録日 | 1994-08-08 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENJ
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | 分子名称: | ENDONUCLEASE V | | 著者 | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | 登録日 | 1994-08-08 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
