5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTY
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8V1I
 
 | |
8V1H
 
 | |
8ZQ3
 
 | |
6WB1
 
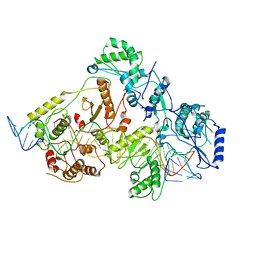 | | +3 extended HIV-1 reverse transcriptase initiation complex core (intermediate state) | | 分子名称: | HIV-1 viral RNA genome fragment, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, reverse transcriptase p51 subunit, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6WBJ
 
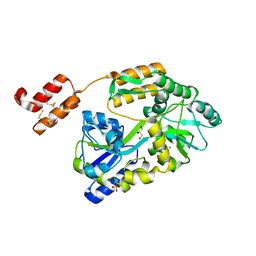 | | High resolution crystal structure of mRECK(CC4) in fusion with engineered MBP | | 分子名称: | GLYCEROL, Maltodextrin-binding protein,Reversion-inducing cysteine-rich protein with Kazal motifs fusion, SULFATE ION, ... | | 著者 | Chang, T.H, Hsieh, F.L, Gabelli, S.B, Nathans, J. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Structure of the RECK CC domain, an evolutionary anomaly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WAZ
 
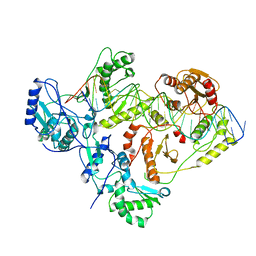 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | 分子名称: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6WBH
 
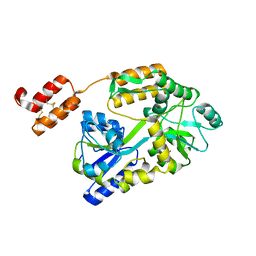 | | Crystal structure of mRECK(CC4) in fusion with engineered MBP at medium resolution | | 分子名称: | CHLORIDE ION, Maltodextrin-binding protein,Reversion-inducing cysteine-rich protein with Kazal motifs fusion, ZINC ION, ... | | 著者 | Chang, T.H, Hsieh, F.L, Gabelli, S.B, Nathans, J. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.455 Å) | | 主引用文献 | Structure of the RECK CC domain, an evolutionary anomaly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W8R
 
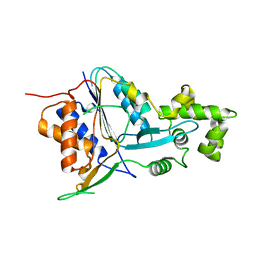 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | 分子名称: | Metacaspase-4, SULFATE ION | | 著者 | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | 登録日 | 2020-03-21 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6WB2
 
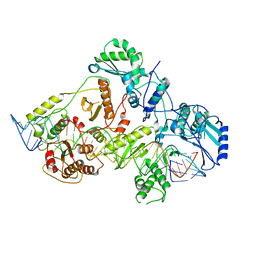 | | +3 extended HIV-1 reverse transcriptase initiation complex core (displaced state) | | 分子名称: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
8A83
 
 | |
8WZQ
 
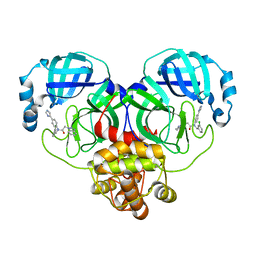 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with CCF0058981 | | 分子名称: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | 著者 | Zou, X.F, Jiang, H.H, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2023-11-02 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WZP
 
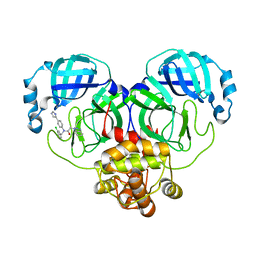 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | 分子名称: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | 著者 | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2023-11-02 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
3ZDA
 
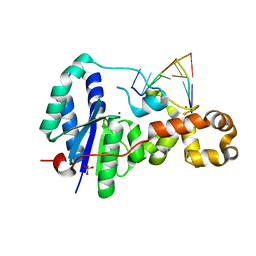 | | Structure of E. coli ExoIX in complex with a fragment of the Flap1 DNA oligonucleotide, potassium and magnesium | | 分子名称: | 5'-D(*AP*AP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP)-3', MAGNESIUM ION, ... | | 著者 | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
8HVY
 
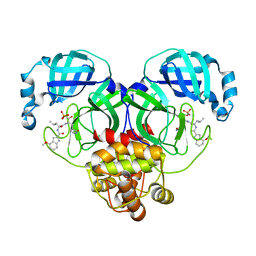 | |
8IGA
 
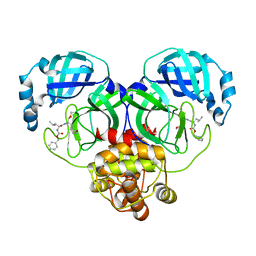 | |
8HVU
 
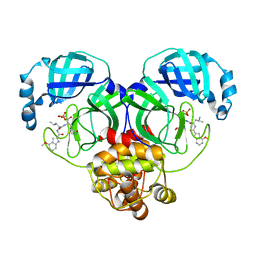 | |
8HVV
 
 | |
8HVX
 
 | |
8HVZ
 
 | |
8HVW
 
 | |
8IG4
 
 | |
8IG5
 
 | | Crystal structure of SARS main protease in complex with GC376 | | 分子名称: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2023-02-20 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | |
