4EW4
 
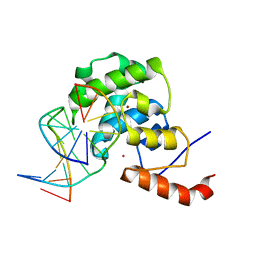 | | mouse MBD4 glycosylase domain in complex with DNA containing a ribose sugar | | 分子名称: | DNA (5'-D(*CP*CP*AP*TP*GP*(3DR)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4, ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2012-04-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.791 Å) | | 主引用文献 | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4F0Q
 
 | | MspJI Restriction Endonuclease - P21 Form | | 分子名称: | MAGNESIUM ION, Restriction endonuclease | | 著者 | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | 登録日 | 2012-05-04 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.046 Å) | | 主引用文献 | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
4F0P
 
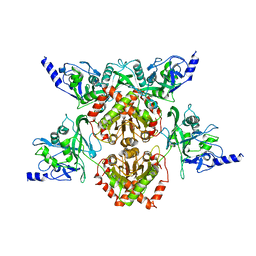 | | MspJI Restriction Endonuclease - P31 Form | | 分子名称: | MAGNESIUM ION, Restriction endonuclease | | 著者 | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | 登録日 | 2012-05-04 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
6VXK
 
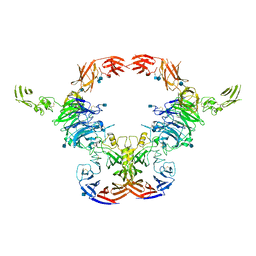 | | Cryo-EM Structure of the full-length A39R/PlexinC1 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-C1, Semaphorin-like protein 139 | | 著者 | Kuo, Y.-C, Chen, H, Shang, G, Uchikawa, E, Tian, H, Bai, X, Zhang, X. | | 登録日 | 2020-02-22 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation.
Nat Commun, 11, 2020
|
|
4FNC
 
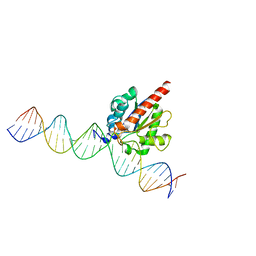 | | Human TDG in a post-reactive complex with 5-hydroxymethyluracil (5hmU) | | 分子名称: | 5-HYDROXYMETHYL URACIL, DNA (28-MER), DNA (29-MER), ... | | 著者 | Hashimoto, H, Hong, S, Bhagwat, A.S, Zhang, X, Cheng, X. | | 登録日 | 2012-06-19 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4GAZ
 
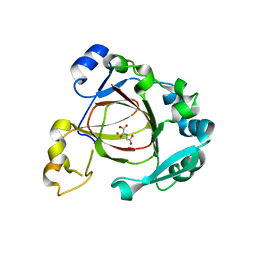 | | Crystal Structure of a Jumonji Domain-containing Protein JMJD5 | | 分子名称: | Lysine-specific demethylase 8, N-OXALYLGLYCINE, NICKEL (II) ION | | 著者 | Wang, H, Zhou, X, Zhang, X, Tao, Y, Chen, N, Zang, J. | | 登録日 | 2012-07-26 | | 公開日 | 2013-08-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Crystal Structure of a Jumonji Domain-containing Protein JMJD5
To be Published
|
|
4GYV
 
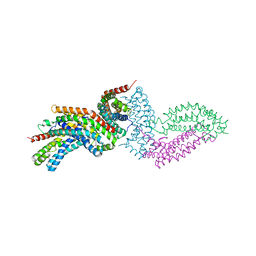 | | Crystal structure of the DH domain of FARP2 | | 分子名称: | FERM, RhoGEF and pleckstrin domain-containing protein 2 | | 著者 | He, X, Zhang, X. | | 登録日 | 2012-09-05 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2.
Structure, 21, 2013
|
|
4GZN
 
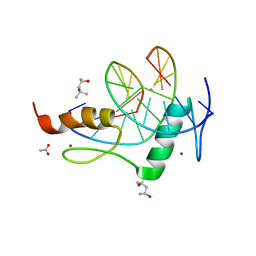 | | Mouse ZFP57 zinc fingers in complex with methylated DNA | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Liu, Y, Zhang, X, Cheng, X. | | 登録日 | 2012-09-06 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence.
Genes Dev., 26, 2012
|
|
4GZU
 
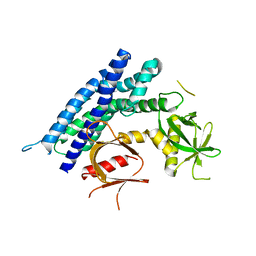 | |
2PJH
 
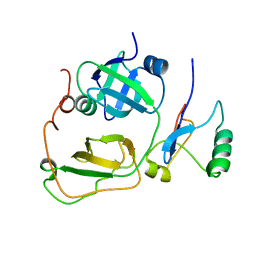 | | Strctural Model of the p97 N domain- npl4 UBD complex | | 分子名称: | Nuclear protein localization protein 4 homolog, Transitional endoplasmic reticulum ATPase | | 著者 | Isaacson, R, Pye, V.E, Simpson, S, Meyer, H.H, Zhang, X, Freemont, P. | | 登録日 | 2007-04-16 | | 公開日 | 2007-05-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Detailed structural insights into the p97-Npl4-Ufd1 interface.
J.Biol.Chem., 282, 2007
|
|
4QWQ
 
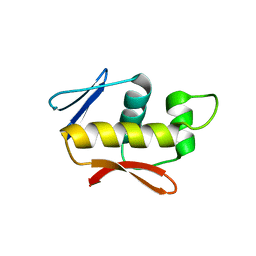 | | Crystal structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus | | 分子名称: | Response regulator SaeR | | 著者 | Fan, X, Zhu, Y, Zhang, X, Teng, M, Li, X. | | 登録日 | 2014-07-17 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3PU8
 
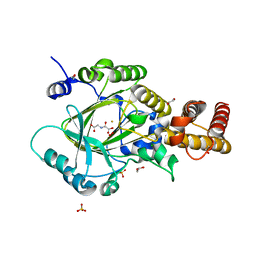 | | PHF2 Jumonji-NOG-Fe(II) complex | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.943 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PUS
 
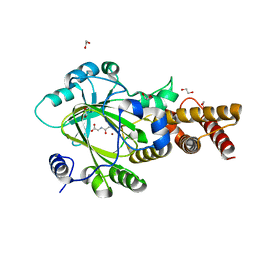 | | PHF2 Jumonji-NOG-Ni(II) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PU3
 
 | | PHF2 Jumonji domain-NOG complex | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PTR
 
 | | PHF2 Jumonji domain | | 分子名称: | 1,2-ETHANEDIOL, PHD finger protein 2 | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PUA
 
 | | PHF2 Jumonji-NOG-Ni(II) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3QXY
 
 | | Human SETD6 in complex with RelA Lys310 | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
3RC0
 
 | | Human SETD6 in complex with RelA Lys310 peptide | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
3QO2
 
 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | 分子名称: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | 著者 | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | 登録日 | 2011-02-09 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
2UXP
 
 | | TtgR in complex Chloramphenicol | | 分子名称: | CHLORAMPHENICOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | 著者 | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | 登録日 | 2007-03-29 | | 公開日 | 2007-05-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
2UXH
 
 | | TtgR in complex with Quercetin | | 分子名称: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | 著者 | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | 登録日 | 2007-03-28 | | 公開日 | 2007-05-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
8I3B
 
 | |
8I3C
 
 | |
8I38
 
 | |
8I3A
 
 | |
