4BV7
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | 分子名称: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | 著者 | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | 登録日 | 2013-06-25 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
6QGB
 
 | | Crystal structure of Ideonella sakaiensis MHETase bound to benzoic acid | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | 著者 | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | 登録日 | 2019-01-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | 分子名称: | GLYCEROL, SAT domain from CazM | | 著者 | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | 登録日 | 2014-10-27 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4BVD
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | 分子名称: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | 著者 | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | 登録日 | 2013-06-25 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4RRO
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | 分子名称: | (4S,4a'R,10a'S)-2-amino-8'-(2-fluoropyridin-3-yl)-1,4a'-dimethyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | 著者 | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | 登録日 | 2014-11-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
1VTD
 
 | | UNUSUAL HELICAL PACKING IN CRYSTALS OF DNA BEARING A MUTATION HOT SPOT | | 分子名称: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*GP*CP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*GP*CP*GP*CP*CP*GP*GP*T)-3') | | 著者 | Timsit, Y, Westhof, E, Fuchs, R.P.P, Moras, D. | | 登録日 | 1996-12-12 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Unusual helical packing in crystals of DNA bearing a mutation hot spot.
Nature, 341, 1989
|
|
3ZP7
 
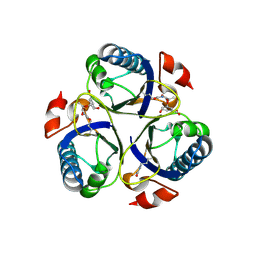 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with chorismate and prephenate | | 分子名称: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, CHORISMATE MUTASE AROH, PREPHENIC ACID | | 著者 | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | 登録日 | 2013-02-26 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZP4
 
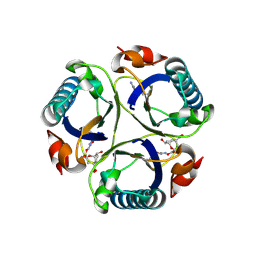 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with a transition state analog | | 分子名称: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE AROH | | 著者 | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | 登録日 | 2013-02-26 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZOP
 
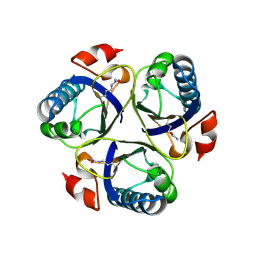 | | Arg90Cit chorismate mutase of Bacillus subtilis at 1.6 A resolution | | 分子名称: | CHORISMATE MUTASE AROH | | 著者 | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | 登録日 | 2013-02-22 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8R7E
 
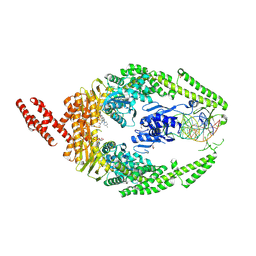 | | MutSbeta bound to compound CHDI-00898647 in the canonical DNA-mismatch bound form | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Abivertinib, ... | | 著者 | Thomsen, M, Thieulin-Pardo, G, Steinbacher, S, Brace, G, Tillack, K, Johnson, P, Ritzefeld, M, Schaertl, S, Frush, E, Warfield, B, Ballantyne, G, Lee, J.-H, Witte, D, Finley, M, Prasad, B, Monteagudo, E, Plotnikov, N, Pacifici, R, Maillard, M, Wilkinson, H, Iyer, R, Dominguez, C, Vogt, T, Felsenfeld, D, Haque, T. | | 登録日 | 2023-11-24 | | 公開日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.783 Å) | | 主引用文献 | Identification of orthosteric inhibitors of MutSbeta ATPase function
Chemrxiv, 2024
|
|
2C2W
 
 | |
3FL3
 
 | | X-ray structure of the ligand free non covalent swapped form of the A19P/Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease | | 分子名称: | Ribonuclease pancreatic, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | 著者 | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | 登録日 | 2008-12-18 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
8R7C
 
 | | MutSbeta bound to compound CHDI-00915542 in the canonical DNA-mismatch bound form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*AP*TP*CP*GP*AP*TP*CP*GP*CP*AP*GP*CP*TP*TP*CP*AP*GP*AP*TP*AP*G)-3'), ... | | 著者 | Thomsen, M, Thieulin-Pardo, G, Steinbacher, S, Brace, G, Tillack, K, Johnson, P, Ritzefeld, M, Schaertl, S, Frush, E, Warfield, B, Ballantyne, G, Lee, J.-H, Witte, D, Finley, M, Prasad, B, Monteagudo, E, Plotnikov, N, Pacifici, R, Maillard, M, Wilkinson, H, Iyer, R, Dominguez, C, Vogt, T, Felsenfeld, D, Haque, T. | | 登録日 | 2023-11-24 | | 公開日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Identification of orthosteric inhibitors of MutSbeta ATPase function
Chemrxiv, 2024
|
|
1FDV
 
 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 MUTANT H221L COMPLEXED WITH NAD+ | | 分子名称: | 17-BETA-HYDROXYSTEROID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | 登録日 | 1998-01-15 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Unusual charge stabilization of NADP+ in 17beta-hydroxysteroid dehydrogenase.
J.Biol.Chem., 273, 1998
|
|
2GPT
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase in complex with tartrate and shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase/ shikimate 5-dehydrogenase, L(+)-TARTARIC ACID, ... | | 著者 | Singh, S.A, Christendat, D. | | 登録日 | 2006-04-18 | | 公開日 | 2006-06-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic channeling in the shikimate pathway
Biochemistry, 45, 2006
|
|
6BW4
 
 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | 分子名称: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | 著者 | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-12-14 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
8R6O
 
 | | Tubulin-4AZA2996 complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Detyrosinated tubulin alpha-1B chain, ... | | 著者 | Herman, J, Vanstreels, E, Bardiot, D, Prota, A.E, Olieric, N, Gao, L.-J, Vercruysse, T, Daems, T, Waer, M, Herdewijn, P, Louat, T, Steinmetz, M.O, De Jonghe, S, Sprangers, B, Daelemans, D. | | 登録日 | 2023-11-22 | | 公開日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 3-nitropyridine analogues as novel microtubule-targeting agents.
Plos One, 19, 2024
|
|
3ZJF
 
 | | A20 OTU domain with irreversibly oxidised Cys103 from 270 min H2O2 soak. | | 分子名称: | A20P50, CHLORIDE ION | | 著者 | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | 登録日 | 2013-01-17 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3FL0
 
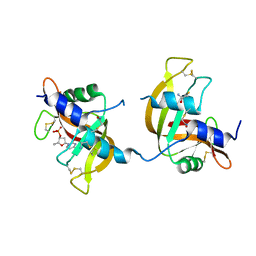 | | X-ray structure of the non covalent swapped form of the Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE | | 分子名称: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, Ribonuclease pancreatic | | 著者 | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | 登録日 | 2008-12-18 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
7TPR
 
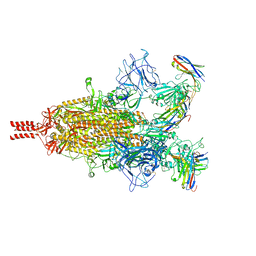 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | 分子名称: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | 著者 | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-20 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
7ZX4
 
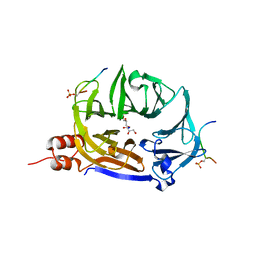 | | Clathrin N-terminal domain in complex with a HURP phospho-peptide | | 分子名称: | CHLORIDE ION, Clathrin heavy chain 1, Disks large-associated protein 5, ... | | 著者 | Kliche, J, Badgujar, D, Dobritzsch, D, Ivarsson, Y. | | 登録日 | 2022-05-20 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Large-scale phosphomimetic screening identifies phospho-modulated motif-based protein interactions.
Mol.Syst.Biol., 19, 2023
|
|
7K4R
 
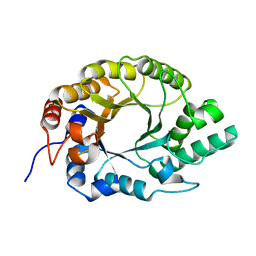 | | Crystal structure of Kemp Eliminase HG3 K50Q | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4T
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (0.999 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4P
 
 | | Crystal structure of Kemp Eliminase HG3 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4S
 
 | | Crystal structure of Kemp Eliminase HG3.7 | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
