8ERQ
 
 | |
8S9G
 
 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | 著者 | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | 登録日 | 2023-03-28 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
6LCV
 
 | |
6LCU
 
 | |
5KYJ
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | 分子名称: | (6~{R})-5-(5-fluoranyl-2-methoxy-pyrimidin-4-yl)-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazole, Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-07-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5KYA
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | 分子名称: | Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta, [2-[(6~{R})-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazol-5-yl]-4-(trifluoromethyl)pyrimidin-5-yl]methanol | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-07-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6U0D
 
 | |
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | 分子名称: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | 著者 | Huang, X. | | 登録日 | 2012-04-20 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | 分子名称: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | 著者 | Huang, X. | | 登録日 | 2012-04-20 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
7LXW
 
 | |
7LY0
 
 | |
7LXZ
 
 | |
7LY2
 
 | |
7LXX
 
 | |
7LXY
 
 | |
7LY3
 
 | |
6MBB
 
 | |
7M10
 
 | |
5DLK
 
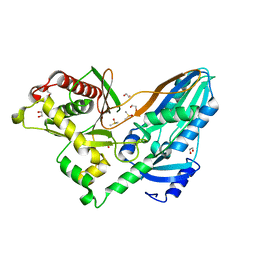 | | The crystal structure of CT mutant | | 分子名称: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, TqaA | | 著者 | Zhang, J.R, Tang, Y, Zhou, J.H. | | 登録日 | 2015-09-06 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
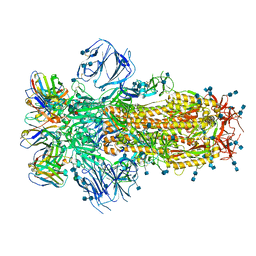
7K43
 
 | |
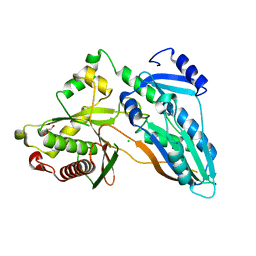
5DIJ
 
 | |
7WH8
 
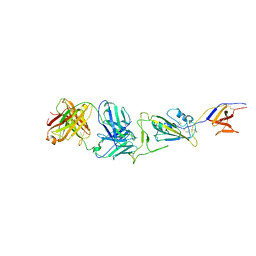 | |
7WHD
 
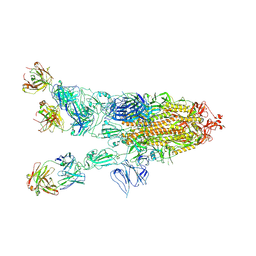 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Zeng, J.W, Wang, X.W, Ge, J.W, Wang, Z.Y. | | 登録日 | 2021-12-30 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
5EJD
 
 | |
5EGF
 
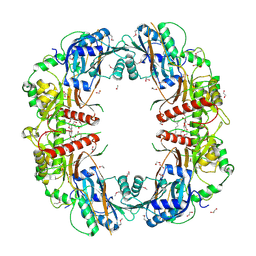 | | The crystal structure of SeMet-CT | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, TqaA | | 著者 | Zhang, J.R, Tang, Y, Zhou, J.H. | | 登録日 | 2015-10-27 | | 公開日 | 2016-10-19 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
