2LN0
 
 | | Structure of MOZ | | 分子名称: | Histone acetyltransferase KAT6A, ZINC ION | | 著者 | Qiu, Y. | | 登録日 | 2011-12-15 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription.
Genes Dev., 26, 2012
|
|
8Y3W
 
 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (same side) | | 分子名称: | DSR anti-defence 1, SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-29 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y34
 
 | | Cryo-EM structure of anti-phage defense associated DSR2 (H171A) (map2) | | 分子名称: | SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-28 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3M
 
 | | Cryo-EM structure of DSR2-DSAD1 complex (cross-linked) | | 分子名称: | DSR anti-defence 1, SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-29 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3Y
 
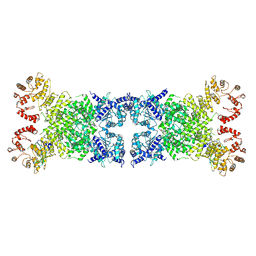 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (opposite side) | | 分子名称: | DSR anti-defence 1, SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-29 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
5T5G
 
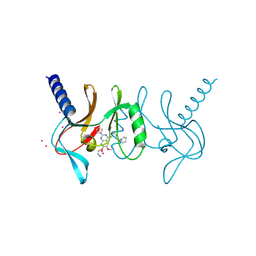 | | human SETD8 in complex with MS2177 | | 分子名称: | 7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)-N-[5-(pyrrolidin-1-yl)pentyl]quinazolin-4-amine, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | 著者 | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-08-30 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
5TH7
 
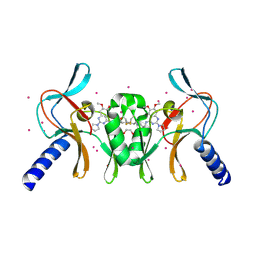 | | Complex of SETD8 with MS453 | | 分子名称: | 1,2-ETHANEDIOL, N-(3-{[6,7-dimethoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)propanamide, N-lysine methyltransferase KMT5A, ... | | 著者 | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-29 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
5YTK
 
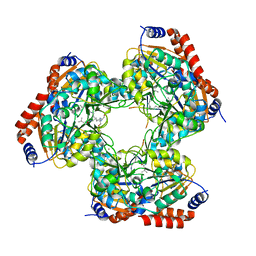 | | Crystal structure of SIRT3 bound to a leucylated AceCS2 | | 分子名称: | AceCS2-KLeu, LEUCINE, LYSINE, ... | | 著者 | Li, J, Gong, W, Xu, Y. | | 登録日 | 2017-11-18 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations
Cell Metab., 27, 2018
|
|
4X0V
 
 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | 分子名称: | Beta-1,3-1,4-glucanase | | 著者 | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | 登録日 | 2014-11-24 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.798 Å) | | 主引用文献 | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
2LNW
 
 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | 分子名称: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | 著者 | Wu, B, Zhang, J, Wu, J, Shi, Y. | | 登録日 | 2012-01-05 | | 公開日 | 2012-11-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LNX
 
 | |
2LUY
 
 | | Solution structure of the tandem zinc finger domain of fission yeast Stc1 | | 分子名称: | Meiotic chromosome segregation protein P8B7.28c, ZINC ION | | 著者 | He, C, Shi, Y, Bayne, E, Wu, J. | | 登録日 | 2012-06-22 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7O2C
 
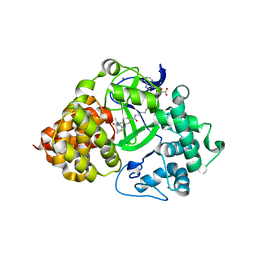 | | X-RAY STRUCTURE OF SMYD3 IN COMPLEX WITH the benzodiazepine-based probe BAY-6035 | | 分子名称: | (2S)-1-[[(1R,5S)-3-azabicyclo[3.1.0]hexan-3-yl]carbonyl]-N-(2-cyclopropylethyl)-2-methyl-4-oxidanylidene-3,5-dihydro-2H-1,5-benzodiazepine-7-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | 著者 | Steuber, H. | | 登録日 | 2021-03-30 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Discovery of the SMYD3 Inhibitor BAY-6035 Using Thermal Shift Assay (TSA)-Based High-Throughput Screening.
Slas Discov, 26, 2021
|
|
7O2B
 
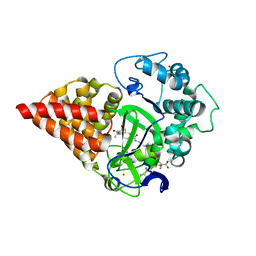 | | X-RAY STRUCTURE OF SMYD3 in complex with benzodiazepine-type inhibitor 6 | | 分子名称: | (2S)-N-butyl-1-(2-fluorophenyl)carbonyl-2-methyl-4-oxidanylidene-3,5-dihydro-2H-1,5-benzodiazepine-7-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | 著者 | Steuber, H. | | 登録日 | 2021-03-30 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery of the SMYD3 Inhibitor BAY-6035 Using Thermal Shift Assay (TSA)-Based High-Throughput Screening.
Slas Discov, 26, 2021
|
|
6KP6
 
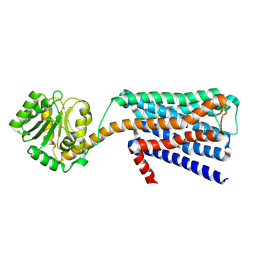 | | The structural study of mutation induced inactivation of Human muscarinic receptor M4 | | 分子名称: | Muscarinic acetylcholine receptor M4,GlgA glycogen synthase,Muscarinic acetylcholine receptor M4 | | 著者 | Wang, J.J, Wu, M, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2019-08-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structural study of mutation-induced inactivation of human muscarinic receptor M4
Iucrj, 7, 2020
|
|
6NM4
 
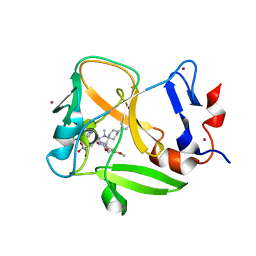 | | Crystal structure of SAM-bound PRDM9 in complex with MRK-740 inhibitor | | 分子名称: | 4-[3-(3,5-dimethoxyphenyl)-1,2,4-oxadiazol-5-yl]-1-methyl-9-(2-methylpyridin-4-yl)-1,4,9-triazaspiro[5.5]undecane, Histone-lysine N-methyltransferase PRDM9, S-ADENOSYLMETHIONINE, ... | | 著者 | Ivanochko, D, Halabelian, L, Fischer, C, Sanders, J.M, Kattar, S.D, Brown, P.J, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2019-01-10 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Discovery of a chemical probe for PRDM9.
Nat Commun, 10, 2019
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | 分子名称: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
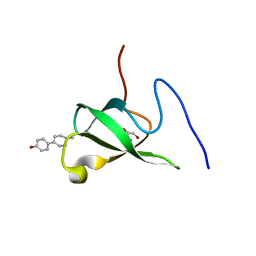 | | Crystal structure of PHF20L1 in complex with Hit 1 | | 分子名称: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-29 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.231 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
3RL0
 
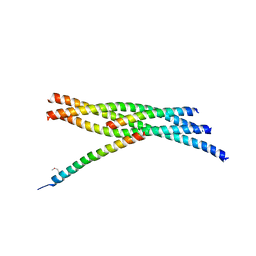 | |
6L1C
 
 | |
6L1I
 
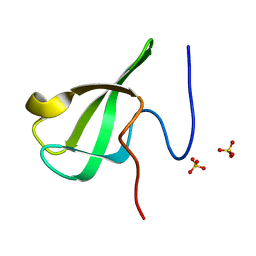 | |
4ER5
 
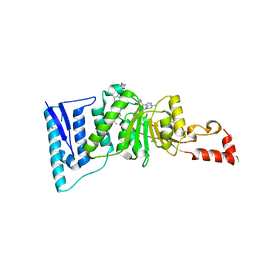 | | Crystal structure of human DOT1L in complex with 2 molecules of EPZ004777 | | 分子名称: | 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | 著者 | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2012-04-19 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
6L10
 
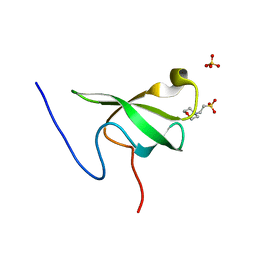 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
3RK3
 
 | |
