8U3H
 
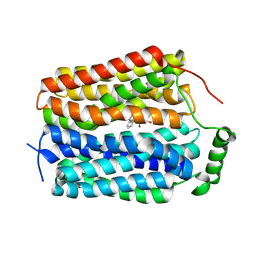 | | Structure of Fmoc-Leu-OH bound Sialin | | 分子名称: | Fluorenylmethyloxycarbonyl chloride, LEUCINE, Sialin | | 著者 | Schmiege, P, Li, X. | | 登録日 | 2023-09-07 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structure and inhibition of the human lysosomal transporter Sialin.
Nat Commun, 15, 2024
|
|
8U3G
 
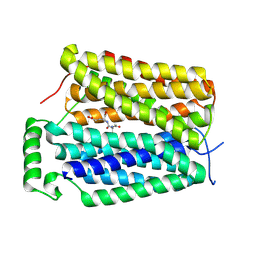 | | Structure of NAAG-bound Sialin | | 分子名称: | ACETYL GROUP, ASPARTIC ACID, GLUTAMIC ACID, ... | | 著者 | Schmiege, P, Li, X. | | 登録日 | 2023-09-07 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Structure and inhibition of the human lysosomal transporter Sialin.
Nat Commun, 15, 2024
|
|
4F8E
 
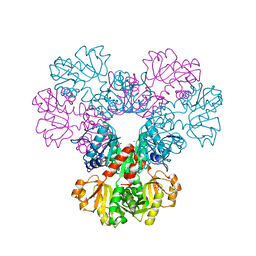 | | Crystal structure of human PRS1 D52H mutant | | 分子名称: | MAGNESIUM ION, Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | 著者 | Chen, P, Teng, M, Li, X. | | 登録日 | 2012-05-17 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | A small disturbance, but a serious disease: the possible mechanism of D52H-mutant of human PRS1 that causes gout
Iubmb Life, 65, 2013
|
|
1QSN
 
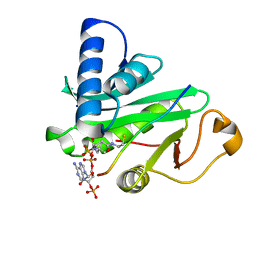 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | 分子名称: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | 著者 | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | 登録日 | 1999-06-22 | | 公開日 | 1999-09-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QXK
 
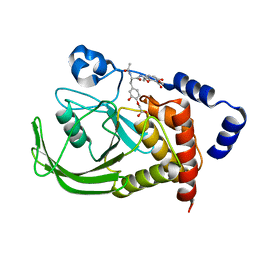 | | Monoacid-Based, Cell Permeable, Selective Inhibitors of Protein Tyrosine Phosphatase 1B | | 分子名称: | 2-{4-[2-ACETYLAMINO-3-(4-CARBOXYMETHOXY-3-HYDROXY-PHENYL)-PROPIONYLAMINO]-BUTOXY}-6-HYDROXY-BENZOIC ACID METHYL ESTER, Protein-tyrosine phosphatase, non-receptor type 1 | | 著者 | Xin, Z, Liu, G, Abad-Zapatero, C, Pei, Z, Szczepankiewick, B.G, Li, X, Zhang, T, Hutchins, C.W, Hajduk, P.J, Ballaron, S.J, Stashko, M.A, Lubben, T.H, Trevillyan, J.M, Jirousek, M.R. | | 登録日 | 2003-09-08 | | 公開日 | 2003-10-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Identification of a Monoacid-Based, Cell Permeable, Selective
Inhibitor of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
4GO5
 
 | |
4GO7
 
 | |
6LAW
 
 | |
6LTR
 
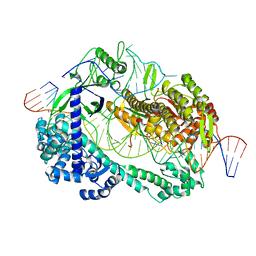 | | Crystal structure of Cas12i2 ternary complex with single Mg2+ bound in catalytic pocket | | 分子名称: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | 著者 | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | 登録日 | 2020-01-23 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LU0
 
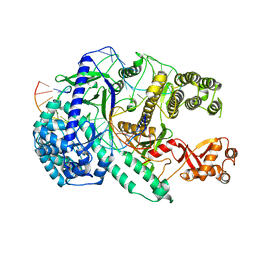 | | Crystal structure of Cas12i2 ternary complex with 12 nt spacer | | 分子名称: | Cas12i2, DNA (5'-D(*GP*CP*CP*GP*CP*TP*TP*TP*CP*TP*T)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*CP*TP*CP*TP*GP*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*C)-3'), ... | | 著者 | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | 登録日 | 2020-01-24 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6P02
 
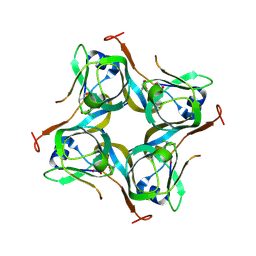 | | Crystal structure of Mtb aspartate decarboxylase, 6-Chlorine pyrazinoic acid complex | | 分子名称: | 6-chloropyrazine-2-carboxylic acid, Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | 著者 | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2019-05-16 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
6OZ8
 
 | |
6LAV
 
 | |
6P1Y
 
 | | Crystal structure of Mtb aspartate decarboxylase mutant M117I | | 分子名称: | AMMONIUM ION, Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, ... | | 著者 | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2019-05-20 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
7SHN
 
 | | Cryo-EM structure of oleoyl-CoA-bound human peroxisomal fatty acid transporter ABCD1 | | 分子名称: | ATP-binding cassette sub-family D member 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | 著者 | Wang, R, Li, X. | | 登録日 | 2021-10-09 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of acyl-CoA transport across the peroxisomal membrane by human ABCD1.
Cell Res., 32, 2022
|
|
7SHM
 
 | |
7TC0
 
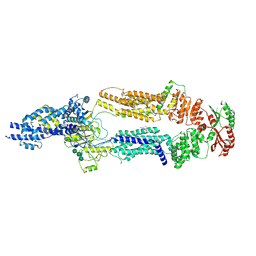 | | The structure of human ABCA1 in digitonin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | 著者 | Sun, Y, Li, X. | | 登録日 | 2021-12-22 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
5ZZ3
 
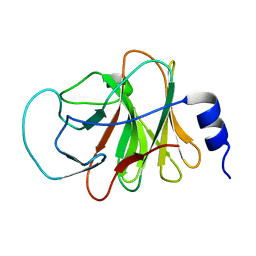 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | 分子名称: | Butyrophilin, subfamily 3, member A3 isoform b variant | | 著者 | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-05-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
7TBY
 
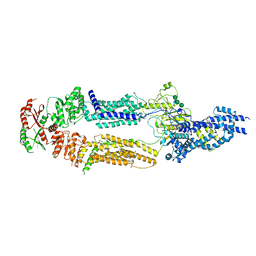 | | The structure of human ABCA1 in nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | 著者 | Sun, Y, Li, X. | | 登録日 | 2021-12-22 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
7TBW
 
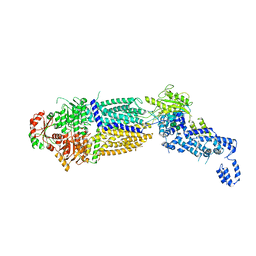 | | The structure of ATP-bound ABCA1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Sun, Y, Li, X. | | 登録日 | 2021-12-22 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
6OHT
 
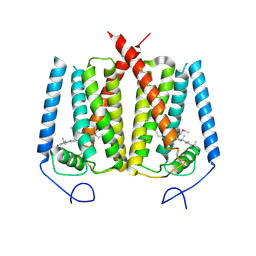 | | Structure of EBP and U18666A | | 分子名称: | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase, 3beta-(2-Diethylaminoethoxy)androst-5-en-17-one | | 著者 | Long, T, Li, X. | | 登録日 | 2019-04-06 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition.
Nat Commun, 10, 2019
|
|
6OYY
 
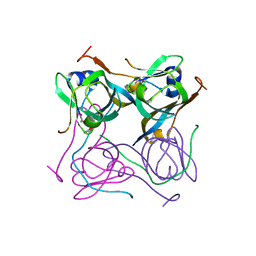 | | Crystal structure of Mtb aspartate decarboxylase, pyrazinoic acid complex | | 分子名称: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, PYRAZINE-2-CARBOXYLIC ACID | | 著者 | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2019-05-15 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
7TBZ
 
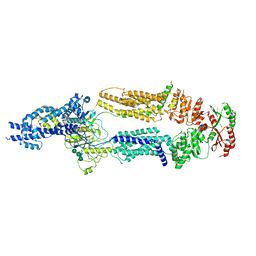 | | The structure of ABCA1 Y482C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | 著者 | Sun, Y, Li, X. | | 登録日 | 2021-12-22 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
6PSL
 
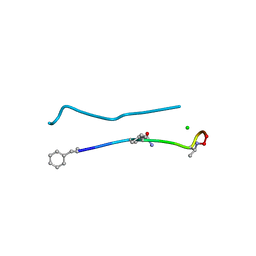 | | Structure of a N-Me-D-Gln4,D-aza-Thr8,Arg10-teixobactin analogue | | 分子名称: | CHLORIDE ION, teixobactin analogue | | 著者 | Nowick, J.S, Yang, H, Pishenko, A, Li, X. | | 登録日 | 2019-07-12 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Design, Synthesis, and Study of Lactam and Ring-Expanded Analogues of Teixobactin.
J.Org.Chem., 85, 2020
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | 分子名称: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | 著者 | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | 登録日 | 2022-12-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
