6IZW
 
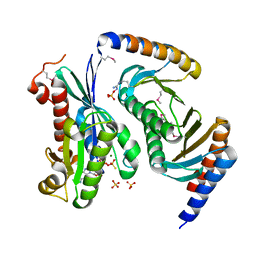 | | Myxococcus xanthus MglA bound to GTP-gamma-S and MglB | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Gliding motility protein MglB, MAGNESIUM ION, ... | | 著者 | Baranwal, J, Gayathri, P. | | 登録日 | 2018-12-20 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Allosteric regulation of a prokaryotic small Ras-like GTPase contributes to cell polarity oscillations in bacterial motility.
Plos Biol., 17, 2019
|
|
4Y1H
 
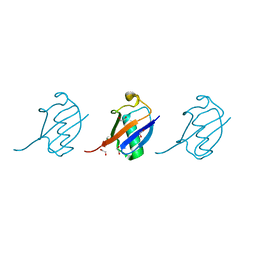 | | Crystal structure of K33 linked tri-Ubiquitin | | 分子名称: | 1,2-ETHANEDIOL, Ubiquitin-40S ribosomal protein S27a | | 著者 | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | 登録日 | 2015-02-07 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
4XYZ
 
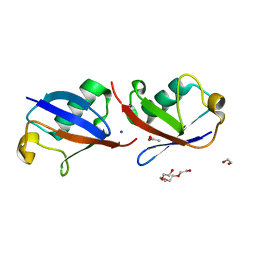 | | Crystal structure of K33 linked di-Ubiquitin | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | 著者 | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | 登録日 | 2015-02-03 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
5KR4
 
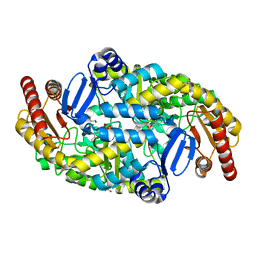 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR6
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-07 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQU
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQT
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminoburyrate transaminase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR3
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQW
 
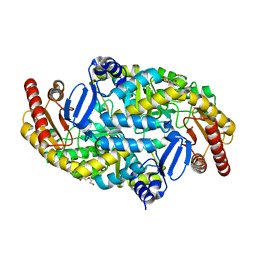 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR5
 
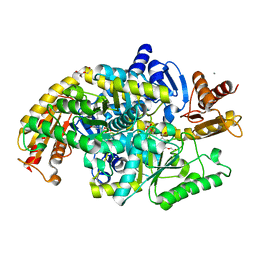 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-07 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
1VGE
 
 | |
6SRR
 
 | |
4XZH
 
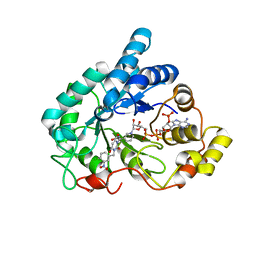 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0048 | | 分子名称: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-chloro-3-nitrobenzyl)-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | 登録日 | 2015-02-04 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZI
 
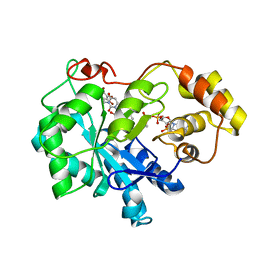 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0049 | | 分子名称: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,4-dioxo-3-(2,3,4,5-tetrabromo-6-methoxybenzyl)-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | 登録日 | 2015-02-04 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZN
 
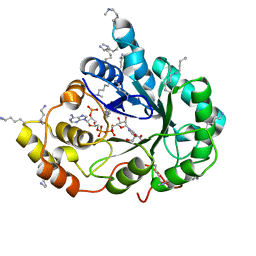 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | 登録日 | 2015-02-04 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZL
 
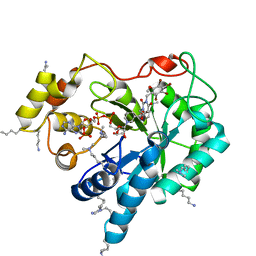 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0049 | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | 登録日 | 2015-02-04 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
6SQU
 
 | | Crystal structure of human SHIP2 catalytic domain in complex with 1,2,4 Dimer | | 分子名称: | 5,5'-(ethane-1,2-diylbis(oxy))bis(benzene-5,4,2,1,-tetrayl)hexakisphosphate, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | 著者 | Whitfield, H, Brearley, C.A, Hemmings, A.M. | | 登録日 | 2019-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention.
J.Med.Chem., 64, 2021
|
|
4XZM
 
 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | 登録日 | 2015-02-04 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4WT2
 
 | | Co-crystal Structure of MDM2 in Complex with AM-7209 | | 分子名称: | 4-({[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetyl}amino)-2-methoxybenzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | 登録日 | 2014-10-30 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of AM-7209, a Potent and Selective 4-Amidobenzoic Acid Inhibitor of the MDM2-p53 Interaction.
J.Med.Chem., 57, 2014
|
|
4YBH
 
 | |
1JZT
 
 | |
1MOR
 
 | |
1MOQ
 
 | |
1RXD
 
 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | 分子名称: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | 著者 | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-12-18 | | 公開日 | 2004-12-28 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
7BKG
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with the tricyclic inhibitor (2) | | 分子名称: | 5,6-dihydro-2-imino-2H,4H-thiazolo(5,4,3-IJ)quinoline, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Schreuder, H.A, Liesum, A. | | 登録日 | 2021-01-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.326 Å) | | 主引用文献 | Novel Inhibitors of Nicotinamide- N -Methyltransferase for the Treatment of Metabolic Disorders.
Molecules, 26, 2021
|
|
