8H57
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) A193P Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-12 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5P
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6N
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (T21I) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-18 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | 分子名称: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H82
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H4Y
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HBK
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M. | | 登録日 | 2022-10-29 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8IJ1
 
 | | Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex | | 分子名称: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | 著者 | Dai, Z, Liang, L, Yin, Y.X. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
5XN3
 
 | |
1NS1
 
 | |
4L08
 
 | | Crystal structure of the maleamate amidase Ami(C149A) in complex with maleate from Pseudomonas putida S16 | | 分子名称: | Hydrolase, isochorismatase family, MALEIC ACID | | 著者 | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | 登録日 | 2013-05-31 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
4L07
 
 | | Crystal structure of the maleamate amidase Ami from Pseudomonas putida S16 | | 分子名称: | GLYCEROL, Hydrolase, isochorismatase family, ... | | 著者 | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | 登録日 | 2013-05-30 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
7W0S
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | 分子名称: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | 著者 | Zhang, H, Liang, X, Li, X.Z. | | 登録日 | 2021-11-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0Q
 
 | |
7W0T
 
 | |
8RKH
 
 | | Crystal structure of the ZP-N2 and ZP-N3 domains of mouse ZP2 (mZP2-N2N3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, ... | | 著者 | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | 登録日 | 2023-12-25 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKE
 
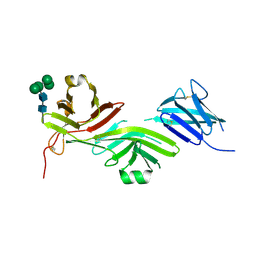 | | Crystal structure of the complete N-terminal region of human ZP2 (hZP2-N1N2N3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | 登録日 | 2023-12-25 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKI
 
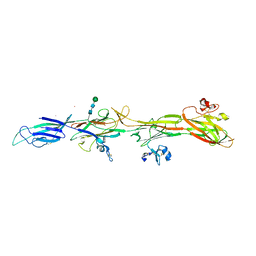 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | 分子名称: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | 著者 | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | 登録日 | 2023-12-25 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKG
 
 | | Crystal structure of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICINE, ... | | 著者 | Nishio, S, de Sanctis, D, Jovine, L. | | 登録日 | 2023-12-25 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKF
 
 | | Crystal structure of the ZP-N1 and ZP-N2 domains of human ZP2 (hZP2-N1N2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2 | | 著者 | Dioguardi, E, Stsiapanava, A, de Sanctis, D, Jovine, L. | | 登録日 | 2023-12-25 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8SK4
 
 | |
8SKH
 
 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent pyrazoline based inhibitors | | 分子名称: | 2-chloro-1-[(4R,5R)-3,4,5-triphenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase nsp5 | | 著者 | Knapp, M.S, Ornelas, E. | | 登録日 | 2023-04-19 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.882 Å) | | 主引用文献 | Discovery of Potent Pyrazoline-Based Covalent SARS-CoV-2 Main Protease Inhibitors.
Chembiochem, 24, 2023
|
|
2B29
 
 | | N-terminal domain of the RPA70 subunit of human replication protein A. | | 分子名称: | Replication protein A 70 kDa DNA-binding subunit | | 著者 | Bochkareva, E, Kaustov, L, Ayed, A, Okorokov, A, Milner, J, Arrowsmith, C.H, Bochkarev, A. | | 登録日 | 2005-09-18 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6L42
 
 | |
2F5J
 
 | |
