6XH7
 
 | | CueR-TAC without RNA | | 分子名称: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Liu, B, Shi, W, Yang, Y. | | 登録日 | 2020-06-18 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH8
 
 | | CueR-transcription activation complex with RNA transcript | | 分子名称: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Liu, B, Shi, W, Yang, Y. | | 登録日 | 2020-06-18 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6A4M
 
 | |
3QE7
 
 | |
5KTX
 
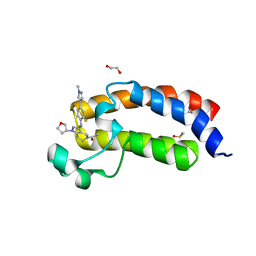 | | CREBBP bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | 分子名称: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, CREB-binding protein, ... | | 著者 | Murray, J.M, Noland, C. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
5KTW
 
 | | CREBBP bromodomain in complex with Cpd 44 (3-((5-acetyl-1-(cyclopropylmethyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl)amino)-N-isopropylbenzamide) | | 分子名称: | 1,2-ETHANEDIOL, 3-[[1-(cyclopropylmethyl)-5-ethanoyl-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]amino]-~{N}-propan-2-yl-benzamide, CREB-binding protein | | 著者 | Murray, J.M, Boenig, G. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.087 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
5KTU
 
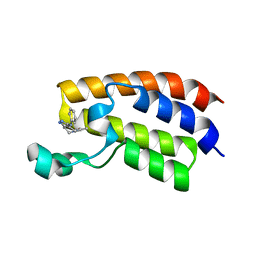 | | Crystal structure of the bromodomain of human CREBBP bound to pyrazolopiperidine scaffold | | 分子名称: | 1-(3-phenylazanyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)ethanone, CREB-binding protein, DIMETHYL SULFOXIDE | | 著者 | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
6WU8
 
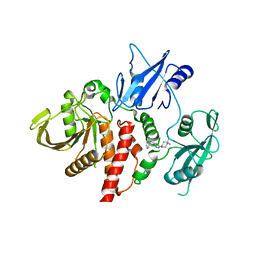 | | Structure of human SHP2 in complex with inhibitor IACS-13909 | | 分子名称: | 1-[3-(2,3-dichlorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Leonard, P.G, Joseph, S, Rodenberger, A. | | 登録日 | 2020-05-04 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Allosteric SHP2 Inhibitor, IACS-13909, Overcomes EGFR-Dependent and EGFR-Independent Resistance Mechanisms toward Osimertinib.
Cancer Res., 80, 2020
|
|
6VN7
 
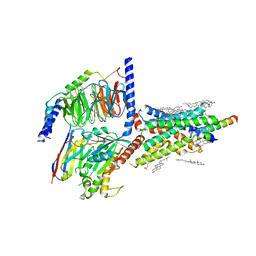 | | Cryo-EM structure of an activated VIP1 receptor-G protein complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Duan, J, Shen, D.-D, Zhou, X.E, Liu, Q.-F, Zhuang, Y.-W, Zhang, H.-B, Xu, P.-Y, Ma, S.-S, He, X.-H, Melcher, K, Zhang, Y, Xu, H.E, Yi, J. | | 登録日 | 2020-01-29 | | 公開日 | 2020-09-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy.
Nat Commun, 11, 2020
|
|
7SAS
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | 著者 | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | 登録日 | 2021-09-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAQ
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | 著者 | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | 登録日 | 2021-09-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAR
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | 著者 | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | 登録日 | 2021-09-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
6PMJ
 
 | |
6PB4
 
 | |
6PB5
 
 | |
6PB6
 
 | |
6PMI
 
 | |
8GTD
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C portal-adaptor complex | | 分子名称: | Head-to-tail joining protein, Portal protein | | 著者 | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTA
 
 | | Cryo-EM structure of the marine siphophage vB_Dshs-R4C capsid | | 分子名称: | Major capsid protein | | 著者 | Sun, H, Huang, Y, Zheng, Q, Li, S, Zhang, R, Xia, N. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTF
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C stopper-terminator complex | | 分子名称: | Head-to-tail joining protein, Major tail protein, Terminator protein | | 著者 | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTB
 
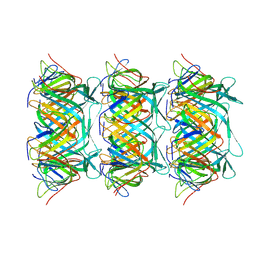 | | Cryo-EM structure of the marine siphophage vB_DshS-R4C tail tube protein | | 分子名称: | Major tail protein | | 著者 | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTC
 
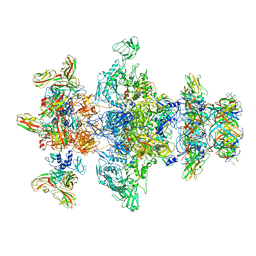 | | Cryo-EM model of the marine siphophage vB_DshS-R4C baseplate-tail complex | | 分子名称: | Distal tail protein, Hub protein, Major tail protein, ... | | 著者 | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
7B1O
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complex with compound 22 | | 分子名称: | 4-chloranyl-N-[(1R)-1-[(1S,5R)-3-quinolin-4-yloxy-6-bicyclo[3.1.0]hexanyl]propyl]benzamide, Indoleamine 2,3-dioxygenase 1 | | 著者 | Lammens, A, Krapp, S, Lewis, R.T, Hamilton, M.M. | | 登録日 | 2020-11-25 | | 公開日 | 2021-09-29 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
6D80
 
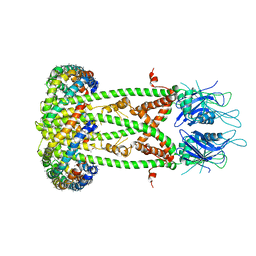 | | Cryo-EM structure of the mitochondrial calcium uniporter from N. fischeri bound to saposin | | 分子名称: | CALCIUM ION, Mitochondrial calcium uniporter, Saposin A | | 著者 | Nguyen, N.X, Armache, J.-P, Cheng, Y, Bai, X.C. | | 登録日 | 2018-04-25 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Cryo-EM structure of a fungal mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | 著者 | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2022-05-11 | | 公開日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
