1EUE
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | 分子名称: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Oganesyan, V, Zhang, X. | | 登録日 | 2000-04-19 | | 公開日 | 2001-04-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5.
FARADAY DISC.CHEM.SOC, 116, 2001
|
|
1F9U
 
 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | 著者 | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | 登録日 | 2000-07-11 | | 公開日 | 2001-06-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1D59
 
 | | CRYSTAL STRUCTURE OF 4-STRANDED OXYTRICHA TELOMERIC DNA | | 分子名称: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | 著者 | Kang, C, Zhang, X, Ratliff, R, Moyzis, R, Rich, A. | | 登録日 | 1992-02-25 | | 公開日 | 1993-04-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of four-stranded Oxytricha telomeric DNA.
Nature, 356, 1992
|
|
1F9W
 
 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | 著者 | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | 登録日 | 2000-07-11 | | 公開日 | 2001-06-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1F9T
 
 | | CRYSTAL STRUCTURES OF KINESIN MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE MOTOR ATPASE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | 著者 | Yun, M, Zhang, X, Park, C.-G, Park, H.-W, Endow, S.A. | | 登録日 | 2000-07-11 | | 公開日 | 2001-06-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1FZX
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAAAAACGG | | 分子名称: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*G)-3' | | 著者 | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | 登録日 | 2000-10-04 | | 公開日 | 2001-03-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
1G14
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAGAAACGG | | 分子名称: | 5'-D(*CP*CP*GP*TP*TP*TP*CP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*GP*AP*AP*AP*CP*GP*G)-3' | | 著者 | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | 登録日 | 2000-10-10 | | 公開日 | 2001-03-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
9L99
 
 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA1-2 | | 分子名称: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | 著者 | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | 登録日 | 2024-12-29 | | 公開日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
9L9A
 
 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA2-3 | | 分子名称: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | 著者 | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | 登録日 | 2024-12-29 | | 公開日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
4ZDS
 
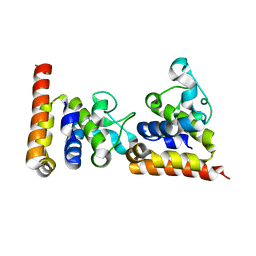 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | 分子名称: | Protein ETHYLENE INSENSITIVE 3 | | 著者 | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | 登録日 | 2015-04-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
5IHW
 
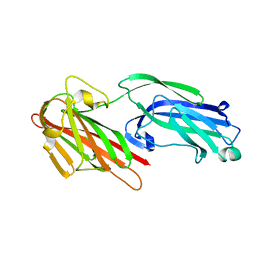 | | The crystal structure of SdrE from staphylococcus aureus | | 分子名称: | Serine-aspartate repeat-containing protein E | | 著者 | Zhang, S, Wei, J, Wu, S, Zhang, X, Luo, M, Wang, D. | | 登録日 | 2016-02-29 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | The crystal structure of SdrE from staphylococcus aureus
To Be Published
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Spacer | | 著者 | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | 登録日 | 2016-02-22 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
5J6P
 
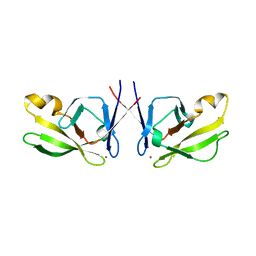 | | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe | | 分子名称: | Kinetochore protein mis18, ZINC ION | | 著者 | Wang, C, Shao, C, Zhang, M, Zhang, X, Zang, J. | | 登録日 | 2016-04-05 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe
To Be Published
|
|
4G1R
 
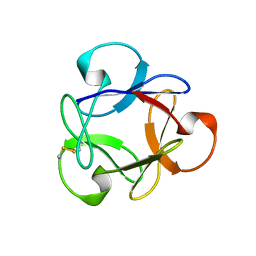 | | Crystal structure of anti-HIV actinohivin in complex with alphs-1,2-mannobiose (Form II) | | 分子名称: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | 著者 | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | 登録日 | 2012-07-11 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Matured structure of anti-HIV lectin actinohivin in complex with alpha-1,2-mannobiose
To be Published
|
|
4END
 
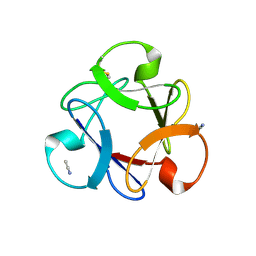 | | Crystal structure of anti-HIV actinohivin in complex with alpha-1,2-mannobiose (P 2 21 21 form) | | 分子名称: | ACETONITRILE, Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | 著者 | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | 登録日 | 2012-04-13 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Matured structure of anti-HIV lectin actinohivin in complex with 1,2-mannobiose
To be Published
|
|
4DEN
 
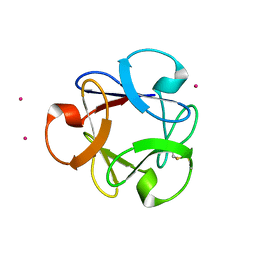 | | Structural insightsinto potent, specific anti-HIV property of actinohivin; Crystal structure of actinohivin in complex with alpha(1-2) mannobiose moiety of high-mannose type glycan of gp120 | | 分子名称: | Actinohivin, POTASSIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | 著者 | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | 登録日 | 2012-01-20 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into the specific anti-HIV property of actinohivin: structure of its complex with the alpha(1–2)mannobiose moiety of gp120
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5CG9
 
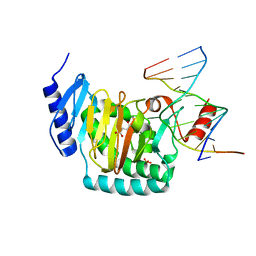 | | NgTET1 in complex with 5mC DNA in space group P3221 | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DNA (5'-D(*TP*GP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | 登録日 | 2015-07-09 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5CG8
 
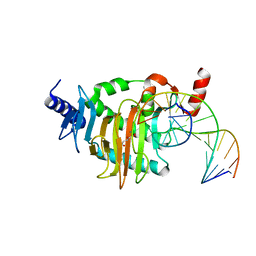 | | NgTET1 in complex with 5hmC DNA | | 分子名称: | 2-OXOGLUTARIC ACID, DNA (5'-D(*AP*GP*AP*AP*TP*TP*CP*CP*GP*TP*TP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*(5HC)P*GP*GP*AP*AP*TP*TP*CP*T)-3'), ... | | 著者 | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | 登録日 | 2015-07-09 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
4G3H
 
 | | Crystal structure of helicobacter pylori arginase | | 分子名称: | Arginase (RocF), MANGANESE (II) ION | | 著者 | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | 登録日 | 2012-07-13 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
3KV9
 
 | | Structure of KIAA1718 Jumonji domain | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVB
 
 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | 分子名称: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV4
 
 | | Structure of PHF8 in complex with histone H3 | | 分子名称: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV5
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
