4UD7
 
 | | Structure of the stapled peptide YS-02 bound to MDM2 | | 分子名称: | MDM2, YS-02 | | 著者 | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | 登録日 | 2014-12-08 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
4UE1
 
 | | Structure of the stapled peptide YS-01 bound to MDM2 | | 分子名称: | E3 UBIQUITIN-PROTEIN LIGASE MDM2, YS-01 | | 著者 | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | 登録日 | 2014-12-14 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
6PCA
 
 | |
6OTA
 
 | |
6PCB
 
 | |
6XVU
 
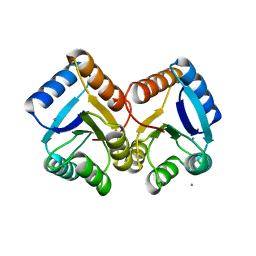 | |
4PXI
 
 | | Elucidation of the Structural and Functional Mechanism of Action of the TetR Family Protein, CprB from S. coelicolor A3(2) | | 分子名称: | CprB, DNA (5'-D(*AP*CP*AP*TP*AP*CP*GP*GP*GP*AP*CP*GP*CP*CP*CP*CP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*CP*GP*GP*GP*GP*CP*GP*TP*CP*CP*CP*GP*TP*AP*TP*GP*T)-3') | | 著者 | Hussain, B, Ruchika, B, Aruna, B, Ruchi, A. | | 登録日 | 2014-03-24 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2)
Nucleic Acids Res., 42, 2014
|
|
5HD4
 
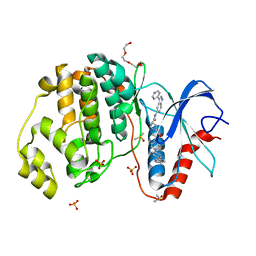 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Wild Type SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | 分子名称: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | 著者 | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | 登録日 | 2016-01-04 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
4OS7
 
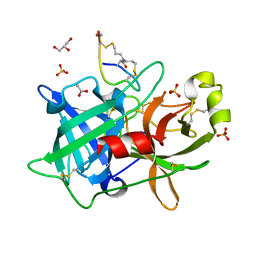 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK607 (bicyclic) | | 分子名称: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Chen, S, Pojer, F, Heinis, C. | | 登録日 | 2014-02-12 | | 公開日 | 2014-09-24 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS5
 
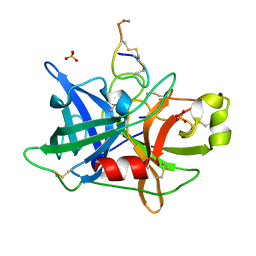 | |
4OS1
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK601 (bicyclic 1) | | 分子名称: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | 著者 | Chen, S, Pojer, F, Heinis, C. | | 登録日 | 2014-02-12 | | 公開日 | 2014-09-24 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
5WO4
 
 | | JAK1 complexed with compound 28 | | 分子名称: | 3-[(4-chloro-3-methoxyphenyl)amino]-1-[(3R,4S)-4-cyanooxan-3-yl]-1H-pyrazole-4-carboxamide, Tyrosine-protein kinase JAK1 | | 著者 | Lesburg, C.A, Patel, S.B. | | 登録日 | 2017-08-01 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | The Discovery of 3-((4-Chloro-3-methoxyphenyl)amino)-1-((3R,4S)-4-cyanotetrahydro-2H-pyran-3-yl)-1H-pyrazole-4-carboxamide, a Highly Ligand Efficient and Efficacious Janus Kinase 1 Selective Inhibitor with Favorable Pharmacokinetic Properties.
J. Med. Chem., 60, 2017
|
|
4OS2
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK602 (bicyclic 1) | | 分子名称: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | 著者 | Chen, S, Pojer, F, Heinis, C. | | 登録日 | 2014-02-12 | | 公開日 | 2014-09-24 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
5USJ
 
 | | Crystal Structure of human KRAS G12D mutant in complex with GDPNP | | 分子名称: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Huang, C.S, Kaplan, A, Stockwell, B.R, Tong, L. | | 登録日 | 2017-02-13 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Multivalent Small-Molecule Pan-RAS Inhibitors.
Cell, 168, 2017
|
|
5HD7
 
 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Mutant SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | 分子名称: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | 登録日 | 2016-01-04 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
4OS6
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK604 (bicyclic 2) | | 分子名称: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | 著者 | Chen, S, Pojer, F, Heinis, C. | | 登録日 | 2014-02-12 | | 公開日 | 2014-09-24 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS4
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK603 (bicyclic 1) | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Chen, S, Pojer, F, Heinis, C. | | 登録日 | 2014-02-12 | | 公開日 | 2014-09-24 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
5ZFO
 
 | |
7N3V
 
 | |
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | 分子名称: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | 著者 | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | 登録日 | 2013-07-19 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4LR6
 
 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | 分子名称: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | 著者 | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | 登録日 | 2013-07-19 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
6QI4
 
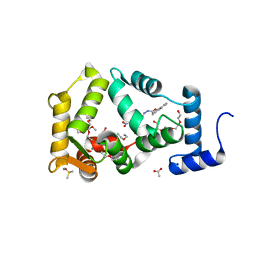 | | NCS-1 bound to a ligand | | 分子名称: | 2-(1~{H}-indol-3-yl)-~{N}-[(~{E})-(4-nitro-3-oxidanyl-phenyl)methylideneamino]ethanamide, ACETATE ION, CALCIUM ION, ... | | 著者 | Sanchez-Barrena, M.J, Blanco-Gabella, P. | | 登録日 | 2019-01-17 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Insights into real-time chemical processes in a calcium sensor protein-directed dynamic library.
Nat Commun, 10, 2019
|
|
7CFD
 
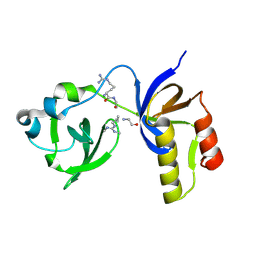 | | Drosophila melanogaster Krimper eTud2-AubR15me2 complex | | 分子名称: | FI20010p1, Protein aubergine | | 著者 | Hu, H, Li, S. | | 登録日 | 2020-06-25 | | 公開日 | 2021-06-02 | | 最終更新日 | 2021-09-15 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFC
 
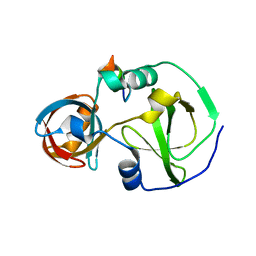 | | Drosophila melanogaster Krimper eTud1-Ago3 complex | | 分子名称: | FI20010p1, Protein argonaute-3 | | 著者 | Hu, H, Li, S. | | 登録日 | 2020-06-25 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFB
 
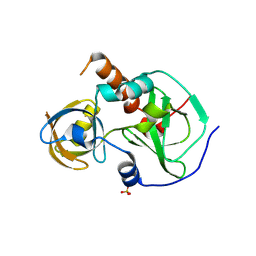 | | Drosophila melanogaster Krimper eTud1 apo structure | | 分子名称: | FI20010p1, SULFATE ION | | 著者 | Hu, H, Li, S. | | 登録日 | 2020-06-25 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
