7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
8JC1
 
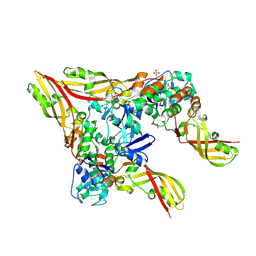 | | Crystal structure of Pectocin M1 from Pectobacterium carotovorum | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | 著者 | Jantarit, N, Kurisu, G, Tanaka, H. | | 登録日 | 2023-05-10 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure of pectocin M1 reveals diverse conformations and interactions during its initial step via the ferredoxin uptake system.
Febs Open Bio, 14, 2024
|
|
7E9O
 
 | |
7E9Q
 
 | |
7E9P
 
 | |
7ENG
 
 | |
7E9N
 
 | |
7ENF
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | 著者 | Wang, X.F, Zhu, Y.Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ERY
 
 | | apo form of the glycosyltransferase | | 分子名称: | Glycosyltransferase | | 著者 | Zhu, X. | | 登録日 | 2021-05-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES2
 
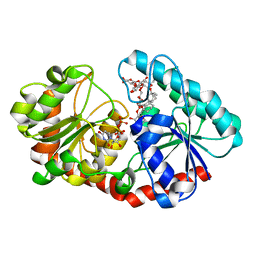 | | a mutant of glycosyktransferase in complex with UDP and Reb D | | 分子名称: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, rebaudioside D | | 著者 | Zhu, X. | | 登録日 | 2021-05-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ERX
 
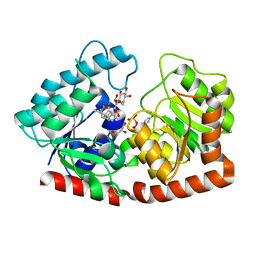 | | Glycosyltransferase in complex with UDP and STB | | 分子名称: | GLYCEROL, Glycosyltransferase, Steviolbioside, ... | | 著者 | Zhu, X. | | 登録日 | 2021-05-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES1
 
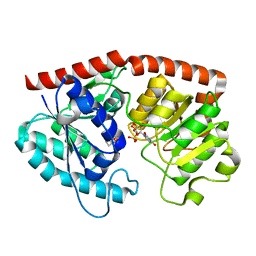 | | glycosyltransferase in complex with UDP and ST | | 分子名称: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, steviol-19-o-glucoside | | 著者 | Zhu, X. | | 登録日 | 2021-05-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES0
 
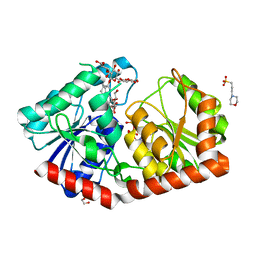 | | a rice glycosyltransferase in complex with UDP and REX | | 分子名称: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | 著者 | Zhu, X. | | 登録日 | 2021-05-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.395 Å) | | 主引用文献 | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
7WX6
 
 | | A Legionella acetyltransferase VipF | | 分子名称: | CHLORAMPHENICOL, COENZYME A, N-acetyltransferase | | 著者 | Chen, T.T, Lin, Y.L, Chen, Z, Han, A.D. | | 登録日 | 2022-02-14 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.273 Å) | | 主引用文献 | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4QR3
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.374 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
7WX7
 
 | | complex of a legionella acetyltransferase VipF and COA/ACO | | 分子名称: | ACETYL COENZYME *A, COENZYME A, N-acetyltransferase | | 著者 | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | 登録日 | 2022-02-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.781 Å) | | 主引用文献 | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WX5
 
 | | a Legionella acetyltransferase effector VipF | | 分子名称: | ACETYL COENZYME *A, N-acetyltransferase | | 著者 | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | 登録日 | 2022-02-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.392 Å) | | 主引用文献 | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | 分子名称: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | 著者 | Guo, M, Fang, P, Wang, J. | | 登録日 | 2020-10-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | 分子名称: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | 著者 | Guo, M, Fang, P, Wang, J. | | 登録日 | 2020-10-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | 分子名称: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | 著者 | Guo, M, Fang, P, Wang, J. | | 登録日 | 2020-10-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | 分子名称: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | 著者 | Li, P, Liu, Z, Fang, P, Wang, J. | | 登録日 | 2021-04-22 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
8YZR
 
 | |
