5Y05
 
 | |
7CAP
 
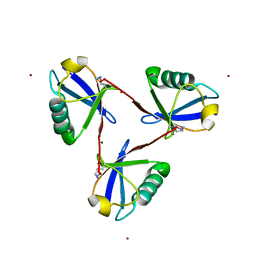 | | Cyclic Lys48-linked triubiquitin | | 分子名称: | Ubiquitin, ZINC ION | | 著者 | Hiranyakorn, M, Yanaka, S, Satoh, T, Wilasri, T, Jityuti, B, Yagi-Utsumi, T, Kato, K. | | 登録日 | 2020-06-09 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 21, 2020
|
|
6K0M
 
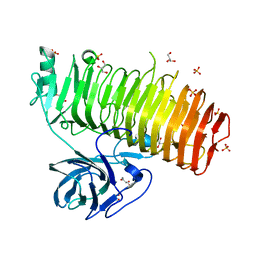 | | Catalytic domain of GH87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11 | | 分子名称: | Alpha-1,3-glucanase, CALCIUM ION, GLYCEROL, ... | | 著者 | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | 登録日 | 2019-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0U
 
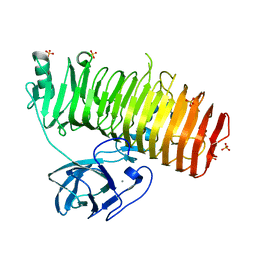 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with tetrasaccharides | | 分子名称: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | 著者 | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | 登録日 | 2019-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
7C7D
 
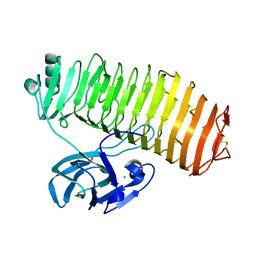 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | 分子名称: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | 著者 | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | 登録日 | 2020-05-25 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
6K0Q
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with nigerose | | 分子名称: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | 著者 | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | 登録日 | 2019-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.564 Å) | | 主引用文献 | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0S
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | 分子名称: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | 著者 | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | 登録日 | 2019-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.534 Å) | | 主引用文献 | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
4NOJ
 
 | | Crystal structure of the mature form of asparaginyl endopeptidase (AEP)/Legumain activated at pH 3.5 | | 分子名称: | Legumain | | 著者 | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | 登録日 | 2013-11-19 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOL
 
 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain mutant D233A at pH 7.5 | | 分子名称: | Legumain | | 著者 | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | 登録日 | 2013-11-19 | | 公開日 | 2014-02-19 | | 最終更新日 | 2014-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOK
 
 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain at pH 7.5 | | 分子名称: | Legumain | | 著者 | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | 登録日 | 2013-11-19 | | 公開日 | 2014-02-19 | | 最終更新日 | 2014-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOM
 
 | | Crystal structure of asparaginyl endopeptidase (AEP)/Legumain activated at pH 4.5 | | 分子名称: | Legumain | | 著者 | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | 登録日 | 2013-11-19 | | 公開日 | 2014-02-19 | | 最終更新日 | 2014-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.006 Å) | | 主引用文献 | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
7Z6P
 
 | |
6K0P
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | 分子名称: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | 著者 | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | 登録日 | 2019-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.424 Å) | | 主引用文献 | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0N
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | 分子名称: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | 著者 | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | 登録日 | 2019-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0V
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | 分子名称: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | 著者 | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | 登録日 | 2019-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.504 Å) | | 主引用文献 | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
7CD9
 
 | | Crystal Structure of SETDB1 tudor domain in complexed with Compound 6 | | 分子名称: | 3-methyl-2-[[(3R,5R)-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, CITRIC ACID, Histone-lysine N-methyltransferase SETDB1 | | 著者 | Xiong, L, Guo, Y, Mao, X, Huang, L, Wu, C, Yang, S. | | 登録日 | 2020-06-19 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7C9N
 
 | | Crystal structure of SETDB1 tudor domain in complexed with Compound 1. | | 分子名称: | 3,5-dimethyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | 著者 | Guo, Y, Xiong, L, Mao, X, Yang, S. | | 登録日 | 2020-06-06 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.472 Å) | | 主引用文献 | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CJT
 
 | | Crystal Structure of SETDB1 Tudor domain in complexed with (R,R)-59 | | 分子名称: | 2-[[(3~{R},5~{R})-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-3-prop-2-enyl-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | 著者 | Guo, Y.P, Liang, X, Mao, X, Wu, C, Luyi, H, Yang, S. | | 登録日 | 2020-07-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.474 Å) | | 主引用文献 | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DMY
 
 | | The crystal structure of Cpd7 in complex with BPTF bromodomain | | 分子名称: | Nucleosome-remodeling factor subunit BPTF, tert-butyl 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-4-oxidanylidene-5,7-dihydropyrrolo[3,4-d]pyrimidine-6-carboxylate | | 著者 | Xiong, L, Guo, Y, Yang, S. | | 登録日 | 2020-12-08 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7DN4
 
 | | The crystal structure of Cpd8 in complex with BPTF bromodomain | | 分子名称: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Xiong, L, Guo, Y, Yang, S. | | 登録日 | 2020-12-08 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.841 Å) | | 主引用文献 | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
1TU3
 
 | | Crystal Structure of Rab5 complex with Rabaptin5 C-terminal Domain | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab GTPase binding effector protein 1, ... | | 著者 | Zhu, G, Zhai, P, Liu, J, Terzyan, S, Li, G, Zhang, X.C. | | 登録日 | 2004-06-24 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural basis of Rab5-Rabaptin5 interaction in endocytosis
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TU4
 
 | | Crystal Structure of Rab5-GDP Complex | | 分子名称: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5A, ... | | 著者 | Zhu, G, Zhai, P, Liu, J, Terzyan, S, Li, G, Zhang, X.C. | | 登録日 | 2004-06-24 | | 公開日 | 2004-10-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of Rab5-Rabaptin5 interaction in endocytosis
Nat.Struct.Mol.Biol., 11, 2004
|
|
2I5Z
 
 | |
3E8N
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) complexed with a potent inhibitor RDEA119 and MgATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | 著者 | Yan, S, Wang, Z.M. | | 登録日 | 2008-08-20 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | RDEA119/BAY 869766: a potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer.
Cancer Res., 69, 2009
|
|
6AQ1
 
 | | The crystal structure of human FABP3 | | 分子名称: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | 著者 | Hsu, H.C, Li, H. | | 登録日 | 2017-08-18 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.40000093 Å) | | 主引用文献 | SAR studies on truxillic acid mono esters as a new class of antinociceptive agents targeting fatty acid binding proteins.
Eur J Med Chem, 154, 2018
|
|
