1Y3I
 
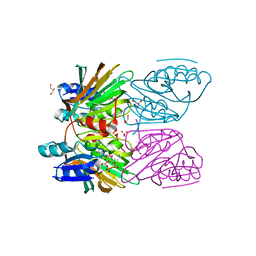 | | Crystal Structure of Mycobacterium tuberculosis NAD kinase-NAD complex | | 分子名称: | GLYCEROL, Inorganic polyphosphate/ATP-NAD kinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, Kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2004-11-25 | | 公開日 | 2005-01-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
Biochem.Biophys.Res.Commun., 327, 2005
|
|
2A4V
 
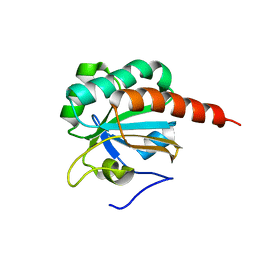 | | Crystal Structure of a truncated mutant of yeast nuclear thiol peroxidase | | 分子名称: | Peroxiredoxin DOT5 | | 著者 | Choi, J, Choi, S, Chon, J.-K, Choi, J, Cha, M.-K, Kim, I.-H, Shin, W. | | 登録日 | 2005-06-29 | | 公開日 | 2006-03-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the C107S/C112S mutant of yeast nuclear 2-Cys peroxiredoxin
Proteins, 61, 2005
|
|
1Y3H
 
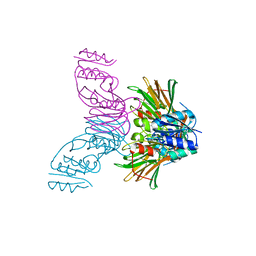 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD kinase from Mycobacterium tuberculosis | | 分子名称: | Inorganic polyphosphate/ATP-NAD kinase | | 著者 | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2004-11-24 | | 公開日 | 2005-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
BIOCHEM.BIOPHYS.RES.COMMUN., 327, 2005
|
|
2AHF
 
 | |
2D8L
 
 | | Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc | | 分子名称: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, Putative glycosyl hydrolase yteR | | 著者 | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2005-12-06 | | 公開日 | 2006-11-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide
J.Mol.Biol., 360, 2006
|
|
2E24
 
 | | crystal structure of a mutant (R612A) of xanthan lyase | | 分子名称: | DI(HYDROXYETHYL)ETHER, Xanthan lyase | | 著者 | Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2006-11-07 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A Structural Factor Responsible for Substrate Recognition by Bacillus sp. GL1 Xanthan Lyase that Acts Specifically on Pyruvated Side Chains of Xanthan
Biochemistry, 46, 2007
|
|
2AHG
 
 | | Unsaturated glucuronyl hydrolase mutant D88N with dGlcA-GalNAc | | 分子名称: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, unsaturated glucuronyl hydrolase | | 著者 | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2005-07-28 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Unsaturated Glucuronyl Hydrolase Complexed with Substrate: MOLECULAR INSIGHTS INTO ITS CATALYTIC REACTION MECHANISM
J.Biol.Chem., 281, 2006
|
|
2CWS
 
 | | Crystal structure at 1.0 A of alginate lyase A1-II', a member of polysaccharide lyase family-7 | | 分子名称: | GLYCEROL, SULFATE ION, alginate lyase A1-II' | | 著者 | Yamasaki, M, Ogura, K, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2005-06-25 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | A Structural Basis for Depolymerization of Alginate by Polysaccharide Lyase Family-7
J.Mol.Biol., 352, 2005
|
|
2D5J
 
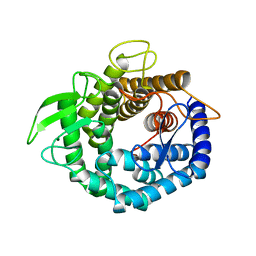 | |
2E22
 
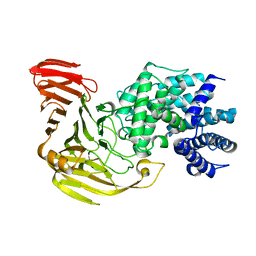 | | Crystal structure of xanthan lyase in complex with mannose | | 分子名称: | Xanthan lyase, alpha-D-mannopyranose | | 著者 | Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2006-11-07 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Structural Factor Responsible for Substrate Recognition by Bacillus sp. GL1 Xanthan Lyase that Acts Specifically on Pyruvated Side Chains of Xanthan
Biochemistry, 46, 2007
|
|
2FUZ
 
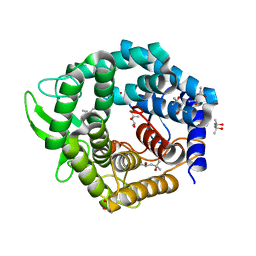 | | UGL hexagonal crystal structure without glycine and DTT molecules | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Unsaturated glucuronyl hydrolase | | 著者 | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2006-01-28 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
7TCQ
 
 | |
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
7N0D
 
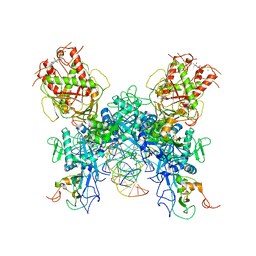 | |
7N0B
 
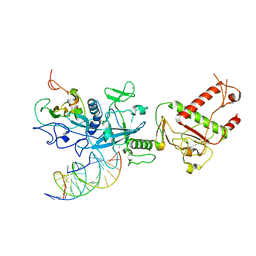 | |
7N0C
 
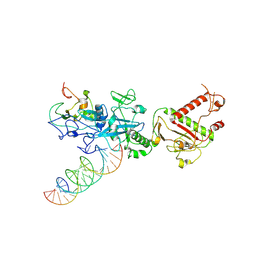 | |
5IQ7
 
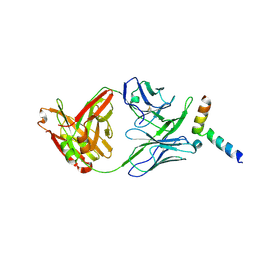 | | Crystal structure of 10E8-S74W Fab in complex with an HIV-1 gp41 peptide. | | 分子名称: | 10E8-S74W Heavy Chain, 10E8-S74W Light Chain, gp41 MPER peptide | | 著者 | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | 登録日 | 2016-03-10 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2869 Å) | | 主引用文献 | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
4Z8M
 
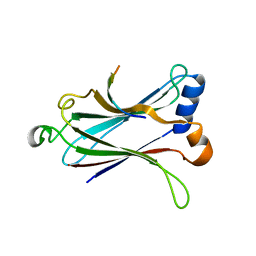 | | Crystal structure of the MAVS-TRAF6 complex | | 分子名称: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | 著者 | Shi, Z.B, Zhou, Z. | | 登録日 | 2015-04-09 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
5VYH
 
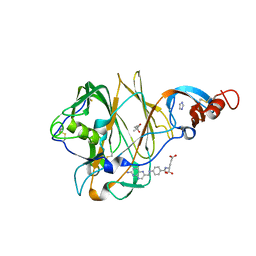 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | 著者 | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | 登録日 | 2017-05-25 | | 公開日 | 2017-08-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6XLK
 
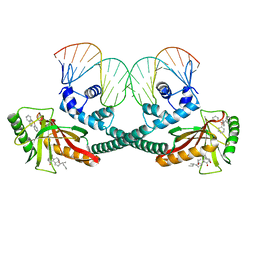 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-4nt | | 分子名称: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | 著者 | Yang, Y, Liu, C, Liu, B. | | 登録日 | 2020-06-28 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLJ
 
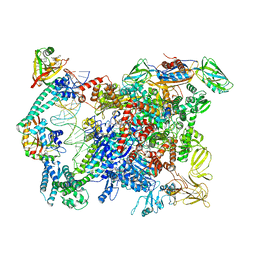 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 4-nt RNA transcript (EcmrR-RPitc-4nt) | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Yang, Y, Liu, C, Liu, B. | | 登録日 | 2020-06-28 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL6
 
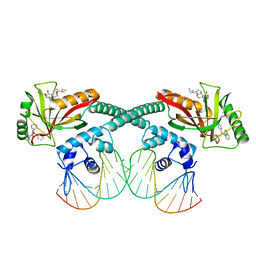 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPo | | 分子名称: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | 著者 | Yang, Y, Liu, C, Liu, B. | | 登録日 | 2020-06-28 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL5
 
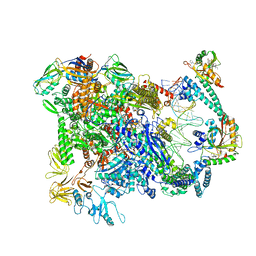 | | Cryo-EM structure of EcmrR-RNAP-promoter open complex (EcmrR-RPo) | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Yang, Y, Liu, C, Liu, B. | | 登録日 | 2020-06-28 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
