7T71
 
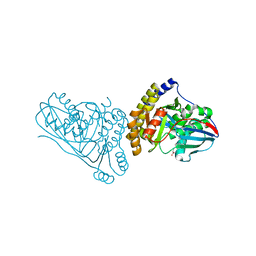 | | Crystal Structure of Mevalonate 3,5-Bisphosphate Decarboxylase from Picrophilus Torridus | | 分子名称: | Mevalonate 3,5-bisphosphate decarboxylase, OLEIC ACID | | 著者 | Vinokur, J.M, Sawaya, M.R, Cascio, D, Collazo, M, Bowie, J.U. | | 登録日 | 2021-12-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways.
J.Biol.Chem., 298, 2022
|
|
6LX4
 
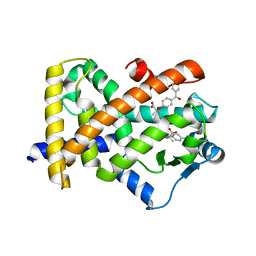 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | 分子名称: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
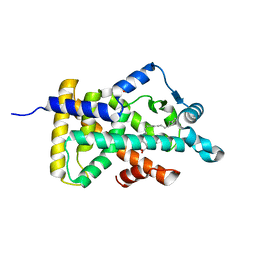 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | 分子名称: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX5
 
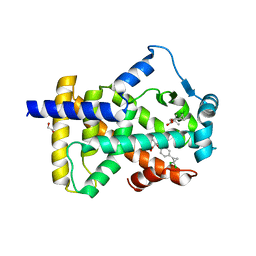 | | X-ray structure of human PPARalpha ligand binding domain-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | 分子名称: | 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX6
 
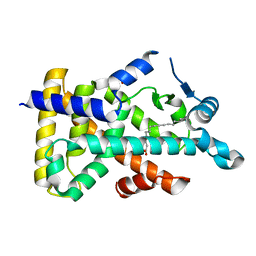 | | X-ray structure of human PPARalpha ligand binding domain-palmitic acid co-crystals obtained by delipidation and cross-seeding | | 分子名称: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
2RQR
 
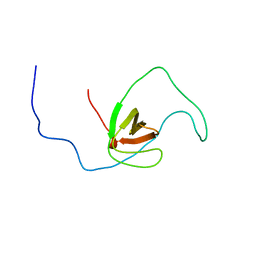 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | 分子名称: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | 著者 | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-10-21 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6LXB
 
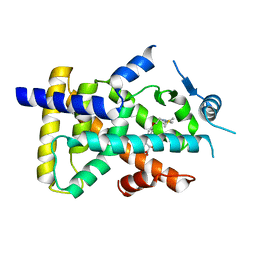 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | 分子名称: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-02-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7EXW
 
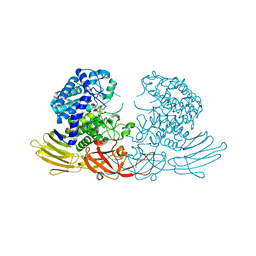 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | 分子名称: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXU
 
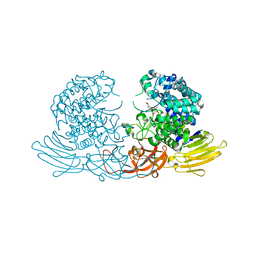 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | 分子名称: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | 分子名称: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
5HXB
 
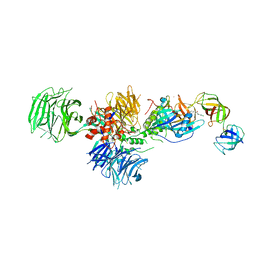 | | Cereblon in complex with DDB1, CC-885, and GSPT1 | | 分子名称: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | 著者 | Chamberlain, P.P, Matyskiela, M, Pagarigan, B. | | 登録日 | 2016-01-30 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Nature, 535, 2016
|
|
3B13
 
 | | Crystal structure of the DHR-2 domain of DOCK2 in complex with Rac1 (T17N mutant) | | 分子名称: | Dedicator of cytokinesis protein 2, Ras-related C3 botulinum toxin substrate 1 | | 著者 | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Mishima-Tsumagari, C, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | 登録日 | 2011-06-24 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.006 Å) | | 主引用文献 | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3A4T
 
 | |
4YOU
 
 | |
4YOT
 
 | |
4YOR
 
 | |
5B5O
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-phenyl-4-((4H-1,2,4-triazol-3-ylsulfanyl)methyl)-1,3-thiazol-2-amine | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | 著者 | Oki, H, Tanaka, Y. | | 登録日 | 2016-05-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
2E9B
 
 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with maltose | | 分子名称: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | 著者 | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | 登録日 | 2007-01-24 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E8Z
 
 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with alpha-cyclodextrin | | 分子名称: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | 著者 | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | 登録日 | 2007-01-24 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
7EAT
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 1 | | 分子名称: | 1,3-dihydro-2H-indol-2-one, SULFATE ION, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, ... | | 著者 | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | 登録日 | 2021-03-08 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBG
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 7 | | 分子名称: | 3,3-dimethyl-7-(methylamino)-1H-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | 登録日 | 2021-03-09 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBB
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 2 | | 分子名称: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | 登録日 | 2021-03-09 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EA0
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 1 | | 分子名称: | 1,3-dihydro-2H-indol-2-one, ACETATE ION, CHLORIDE ION, ... | | 著者 | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | 登録日 | 2021-03-05 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBH
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 13 | | 分子名称: | 5-bromanyl-2-methyl-6-propyl-7H-pyrrolo[2,3-d]pyrimidine, ACETATE ION, CHLORIDE ION, ... | | 著者 | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | 登録日 | 2021-03-09 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EAS
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 2 | | 分子名称: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, ACETATE ION, CHLORIDE ION, ... | | 著者 | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | 登録日 | 2021-03-08 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
