4WT2
 
 | | Co-crystal Structure of MDM2 in Complex with AM-7209 | | 分子名称: | 4-({[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetyl}amino)-2-methoxybenzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | 登録日 | 2014-10-30 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of AM-7209, a Potent and Selective 4-Amidobenzoic Acid Inhibitor of the MDM2-p53 Interaction.
J.Med.Chem., 57, 2014
|
|
8YSA
 
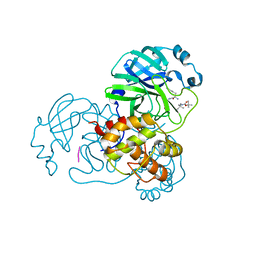 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H102 | | 分子名称: | 3C-like proteinase nsp5, BOC-TBG-PHE-ELL | | 著者 | Zheng, W.Y, Fu, L.F, Feng, Y, Han, P, Qi, J.X. | | 登録日 | 2024-03-22 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery, Biological Activity, and Structural Mechanism of a Potent Inhibitor of SARS-CoV-2 Main Protease
To Be Published
|
|
8VXA
 
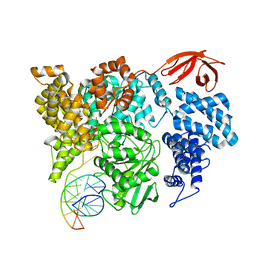 | |
8VX9
 
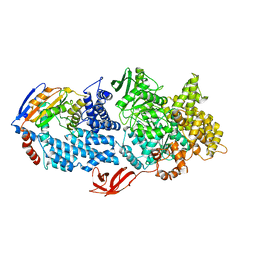 | |
8VXC
 
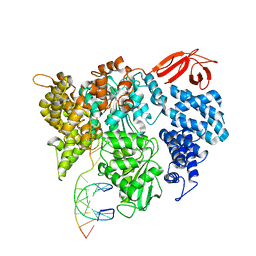 | |
8VXY
 
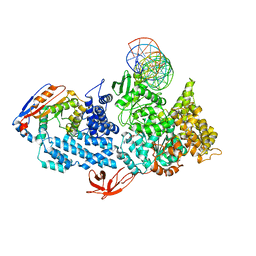 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | 著者 | Tuck, O.T, Hu, J.J, Doudna, J.A. | | 登録日 | 2024-02-06 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | 著者 | He, Q.W, Xu, Z.P, Xie, Y.F. | | 登録日 | 2023-01-02 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
8HWT
 
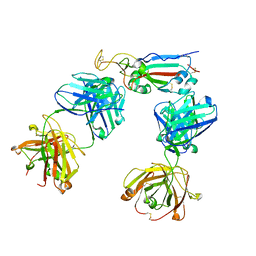 | |
8I1V
 
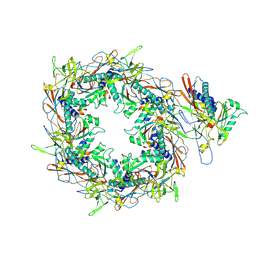 | | The asymmetric unit of P22 procapsid | | 分子名称: | Major capsid protein, Scaffolding protein | | 著者 | Xiao, H, Liu, H.R, Cheng, L.P. | | 登録日 | 2023-01-13 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8I1T
 
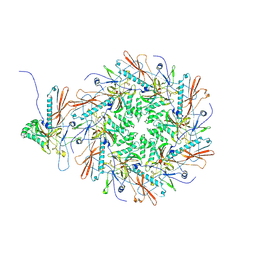 | |
8I99
 
 | | N-carbamoyl-D-amino-acid hydrolase mutant - M4Th3 | | 分子名称: | N-carbamoyl-D-amino-acid hydrolase | | 著者 | Hu, J.M, Ni, Y, Xu, G.C. | | 登録日 | 2023-02-06 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Engineering the Thermostability of a d-Carbamoylase Based on Ancestral Sequence Reconstruction for the Efficient Synthesis of d-Tryptophan.
J.Agric.Food Chem., 71, 2023
|
|
3WY7
 
 | | Crystal structure of Mycobacterium smegmatis 7-Keto-8-aminopelargonic acid (KAPA) synthase BioF | | 分子名称: | 8-amino-7-oxononanoate synthase | | 著者 | Fan, S.H, Li, D.F, Wang, D.C, Chen, G.J, Zhang, X.E, Bi, L.J. | | 登録日 | 2014-08-20 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase
Int.J.Biochem.Cell Biol., 58C, 2014
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | 分子名称: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | 著者 | Hong, W, Chen, H, Wu, Q, Lin, T. | | 登録日 | 2019-09-23 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.45 Å) | | 主引用文献 | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YJ5
 
 | |
8SX3
 
 | |
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | 著者 | Wang, K, Wang, X, Pan, L. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2022-08-18 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2022-08-19 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6Y9X
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,7) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Z
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Y
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-7,13) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | 分子名称: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-06-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
