4XR9
 
 | | Crystal structure of CalS8 from Micromonospora echinospora cocrystallized with NAD and TDP-glucose | | 分子名称: | CalS8, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-01-20 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of CalS8 from Micromonospora echinospora
To Be Published
|
|
4X2R
 
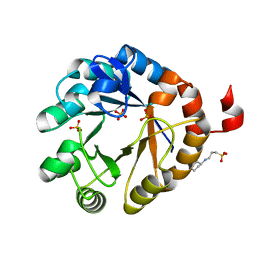 | | Crystal structure of PriA from Actinomyces urogenitalis | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION | | 著者 | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-26 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Evolution of substrate specificity in a retained enzyme driven by gene loss.
Elife, 6, 2017
|
|
6W1I
 
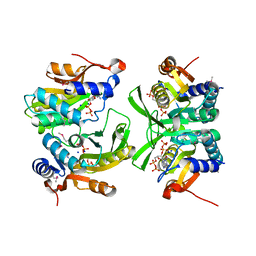 | | Re-interpretation of ppGpp (G4P) electron density in the deposited crystal structure of Xanthine phosphoribosyltransferase (XPRT) (1Y0B). | | 分子名称: | GUANOSINE-5',3'-TETRAPHOSPHATE, SODIUM ION, Xanthine phosphoribosyltransferase | | 著者 | Satyshur, K.A, Anderson, B.W, Keck, J.L, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2020-03-04 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular Mechanism of Regulation of the Purine Salvage Enzyme XPRT by the Alarmones pppGpp, ppGpp, and pGpp.
J.Mol.Biol., 432, 2020
|
|
3FZ4
 
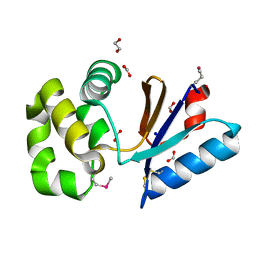 | | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159 | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, Putative arsenate reductase, ... | | 著者 | Tan, K, Hatzos, C, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-01-23 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159.
To be Published
|
|
4WRP
 
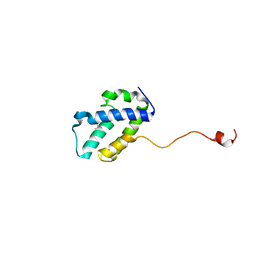 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | 分子名称: | Uncharacterized protein | | 著者 | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-24 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
3E0K
 
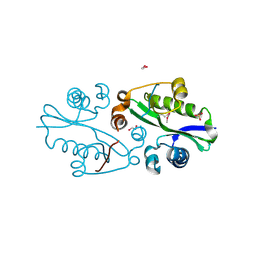 | |
4XI1
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, wild-type | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, GLYCEROL, ... | | 著者 | Stogios, P.J, Quaile, T, Skarina, T, Cuff, M, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-01-06 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.983 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
3DSB
 
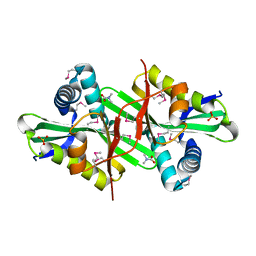 | |
4YE5
 
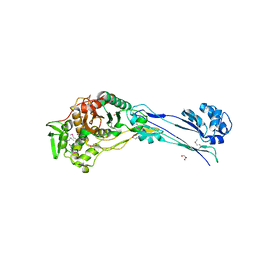 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | 分子名称: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | 著者 | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-02-23 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.052 Å) | | 主引用文献 | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
4WZ2
 
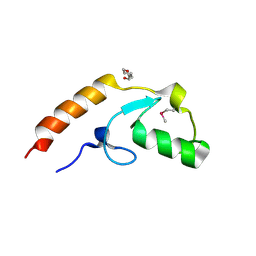 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | 著者 | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.408 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
5I4Q
 
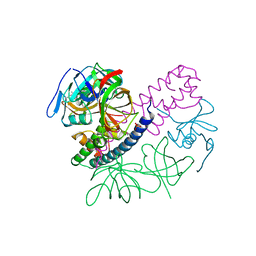 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | 分子名称: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-02-12 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
3FEU
 
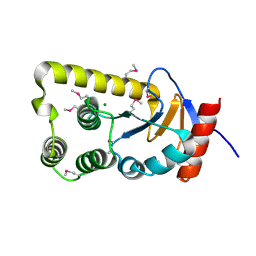 | |
4WZ3
 
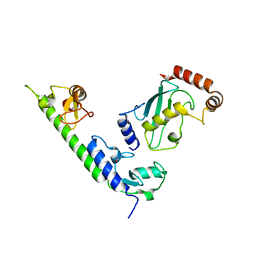 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | 分子名称: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | 著者 | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-07 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
5I4R
 
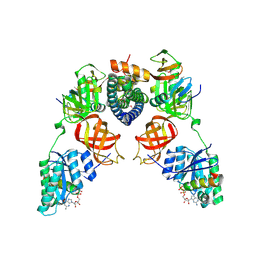 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | 分子名称: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-02-12 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
3FK8
 
 | |
4WUI
 
 | | Crystal structure of TrpF from Jonesia denitrificans | | 分子名称: | CITRIC ACID, N-(5'-phosphoribosyl)anthranilate isomerase | | 著者 | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-31 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4XTK
 
 | | Structure of TM1797, a CAS1 protein from Thermotoga maritima | | 分子名称: | CRISPR-associated endonuclease Cas1 | | 著者 | Petit, P, Beloglazova, N, Skarina, T, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-01-23 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure and nuclease activity of tm1797, a cas1 protein from thermotoga maritima
To Be Published
|
|
4Y7D
 
 | | Alpha/beta hydrolase fold protein from Nakamurella multipartita | | 分子名称: | Alpha/beta hydrolase fold protein, CHLORIDE ION, SODIUM ION | | 著者 | Cuff, M.E, OSIPIUK, J, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-02-14 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Alpha/beta hydrolase fold protein from Nakamurella multipartita.
to be published
|
|
4WZ0
 
 | | Crystal structure of U-box 1 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris | | 分子名称: | E3 ubiquitin-protein ligase LubX | | 著者 | Stogios, P.J, Quaile, A.T, Skarina, T, Stein, A, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
3F6W
 
 | | XRE-family like protein from Pseudomonas syringae pv. tomato str. DC3000 | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, XRE-family like protein | | 著者 | Petrova, T, Cuff, M, Shackleford, G, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-11-06 | | 公開日 | 2008-12-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | XRE-family like protein from Pseudomonas syringae pv. tomato str. DC3000
To be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | 著者 | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-09-13 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
3DCA
 
 | | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target | | 分子名称: | RPA0582, SULFATE ION | | 著者 | Sledz, P, Wang, S, Chruszcz, M, Yim, V, Kudritska, M, Evdokimova, E, Turk, D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-06-03 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target
To be Published
|
|
4XLT
 
 | | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053 | | 分子名称: | Response regulator receiver protein | | 著者 | Chang, C, Cuff, M, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-01-13 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053
To Be Published
|
|
5IOB
 
 | | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase-related glycosidases, CHLORIDE ION, ... | | 著者 | Chang, C, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-03-08 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum
To Be Published
|
|
5HKQ
 
 | | Crystal structure of CDI complex from Escherichia coli STEC_O31 | | 分子名称: | CdiI immunity protein, Contact-dependent inhibitor A | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional plasticity of antibacterial EndoU toxins.
Mol.Microbiol., 109, 2018
|
|
