1TYF
 
 | |
7LG7
 
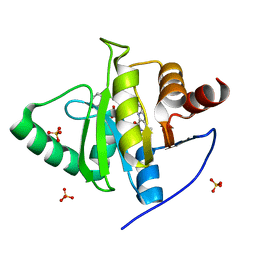 | | Crystal structure of CoV-2 Nsp3 Macrodomain complex with PARG345 | | 分子名称: | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide, Non-structural protein 3, SULFATE ION | | 著者 | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | 登録日 | 2021-01-19 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
3UPI
 
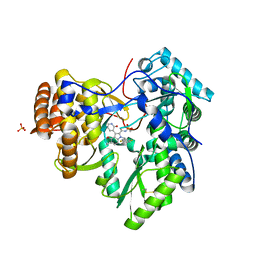 | | Synthesis of novel 4,5-dihydrofurano indoles and their evaluation as HCV NS5B polymerase inhibitors | | 分子名称: | (3S)-6-(2,5-difluorobenzyl)-3-methyl-N-(methylsulfonyl)-8-(2-oxo-1,2-dihydropyridin-3-yl)-3,6-dihydro-2H-furo[2,3-e]indole-7-carboxamide, PHOSPHATE ION, RNA-directed RNA polymerase | | 著者 | Velazquez, F, Venkataraman, S, Lesburg, C.A, Duca, J.S, Rosenblum, S.B, Kozlowski, J.A, Njoroge, F.G. | | 登録日 | 2011-11-18 | | 公開日 | 2012-01-25 | | 最終更新日 | 2012-02-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Synthesis of New 4,5-Dihydrofuranoindoles and Their Evaluation as HCV NS5B Polymerase Inhibitors.
Org.Lett., 14, 2012
|
|
3FWH
 
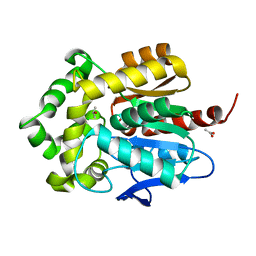 | | Structure of haloalkane dehalogenase mutant Dha15 (I135F/C176Y) from Rhodococcus rhodochrous | | 分子名称: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | 著者 | Gavira, J.A, Stsiapanava, A, Kuty, M, Dohnalek, J, Lapkouski, M, Kuta Smatanova, I. | | 登録日 | 2009-01-18 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3FY4
 
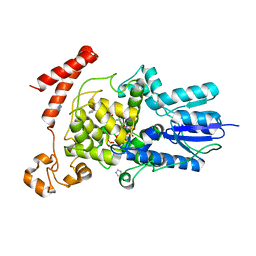 | | (6-4) Photolyase Crystal Structure | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-4 photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Hitomi, K, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2009-01-21 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5D5L
 
 | |
1RXV
 
 | | Crystal Structure of A. Fulgidus FEN-1 bound to DNA | | 分子名称: | 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG), Flap structure-specific endonuclease | | 著者 | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | 登録日 | 2003-12-18 | | 公開日 | 2004-01-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
3UXW
 
 | | Crystal Structures of an A-T-hook/DNA complex | | 分子名称: | A-T hook peptide, dodecamer DNA | | 著者 | Fonfria-Subiros, E, Acosta-Reyes, F.J, Saperas, N, Pous, J, Subirana, J.A, Campos, J.L. | | 登録日 | 2011-12-05 | | 公開日 | 2012-05-23 | | 最終更新日 | 2013-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal structure of a complex of DNA with one AT-hook of HMGA1.
Plos One, 7, 2012
|
|
7JT4
 
 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | 分子名称: | Nucleosome-remodeling factor subunit BPTF | | 著者 | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2020-08-17 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
3G1H
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate | | 分子名称: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-01-29 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
1S23
 
 | | Crystal Structure Analysis of the B-DNA Decamer CGCAATTGCG | | 分子名称: | 5'-D(*CP*GP*CP*AP*AP*TP*TP*GP*CP*G)-3', COBALT (II) ION | | 著者 | Valls, N, Wright, G, Steiner, R.A, Murshudov, G.N, Subirana, J.A. | | 登録日 | 2004-01-08 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | DNA variability in five crystal structures of d(CGCAATTGCG).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5A4C
 
 | | FGFR1 ligand complex | | 分子名称: | 1,2-ETHANEDIOL, 1-tert-butyl-3-[2-[3-(diethylamino)propylamino]-6-(3,5-dimethoxyphenyl)pyrido[2,3-d]pyrimidin-7-yl]urea, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | 著者 | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | 登録日 | 2015-06-05 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
7JRS
 
 | |
3V5C
 
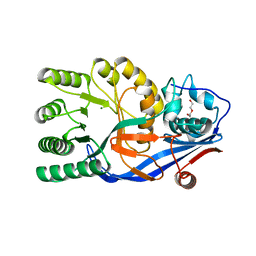 | | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | 著者 | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-12-16 | | 公開日 | 2012-12-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg
To be Published
|
|
1S5T
 
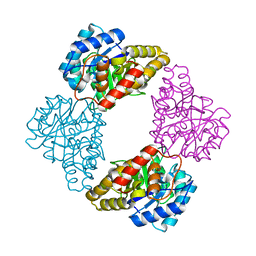 | |
3ZE5
 
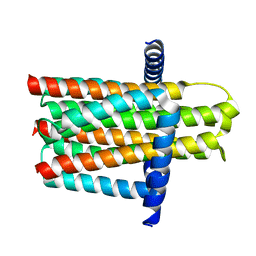 | | Crystal structure of the integral membrane diacylglycerol kinase - delta4 | | 分子名称: | DIACYLGLYCEROL KINASE | | 著者 | Li, D, Vogeley, L, Pye, V.E, Lyons, J.A, Aragao, D, Caffrey, M. | | 登録日 | 2012-12-03 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
3ZE3
 
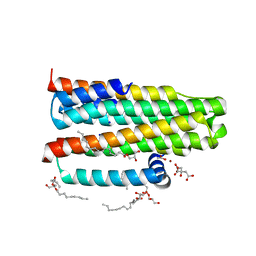 | | Crystal structure of the integral membrane diacylglycerol kinase - delta7 | | 分子名称: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | 著者 | Li, D, Pye, V.E, Lyons, J.A, Vogeley, L, Aragao, D, Caffrey, M. | | 登録日 | 2012-12-03 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
3DZ6
 
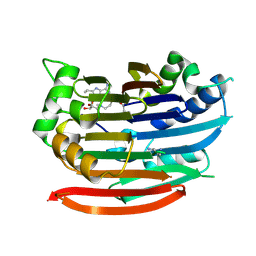 | | Human AdoMetDC with 5'-[(4-aminooxybutyl)methylamino]-5'deoxy-8-ethyladenosine | | 分子名称: | 1,4-DIAMINOBUTANE, 5'-{[4-(aminooxy)butyl](methyl)amino}-5'-deoxy-8-ethenyladenosine, S-adenosylmethionine decarboxylase alpha chain, ... | | 著者 | Bale, S, McCloskey, D.E, Pegg, A.E, Secrist III, J.A, Guida, W.C, Ealick, S.E. | | 登録日 | 2008-07-29 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | New Insights into the Design of Inhibitors of Human S-Adenosylmethionine Decarboxylase: Studies of Adenine C8 Substitution in Structural Analogues of S-Adenosylmethionine
J.Med.Chem., 52, 2009
|
|
3ZE4
 
 | | Crystal structure of the integral membrane diacylglycerol kinase - wild-type | | 分子名称: | DIACYLGLYCEROL KINASE | | 著者 | Li, D, Lyons, J.A, Pye, V.E, Vogeley, L, Aragao, D, Caffrey, M. | | 登録日 | 2012-12-03 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.702 Å) | | 主引用文献 | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
7JO9
 
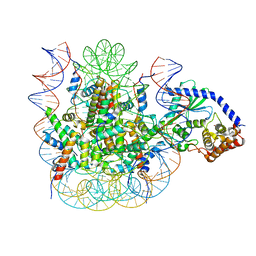 | | 1:1 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | 著者 | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | 登録日 | 2020-08-06 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
1SH2
 
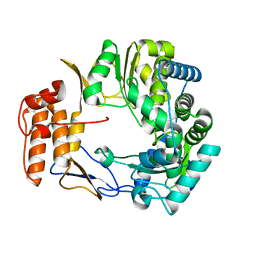 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | 分子名称: | RNA Polymerase | | 著者 | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | 登録日 | 2004-02-24 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1SFF
 
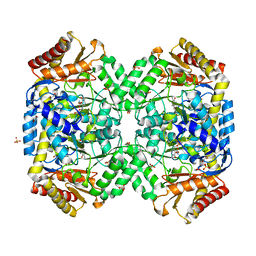 | | Structure of gamma-aminobutyrate aminotransferase complex with aminooxyacetate | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | 著者 | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-02-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
3ZC3
 
 | | FERREDOXIN-NADP REDUCTASE (MUTATION S80A) COMPLEXED WITH NADP BY COCRYSTALLIZATION | | 分子名称: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Martinez-Julvez, M, Hurtado-Guerrero, R, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | 登録日 | 2012-11-15 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
3ZDJ
 
 | |
3ZIV
 
 | |
