7YFL
 
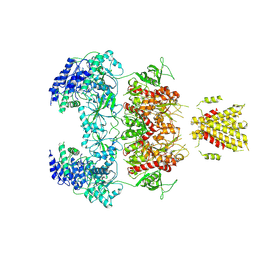 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, J.L, Zhu, S.J, Zhang, M. | | 登録日 | 2022-07-08 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
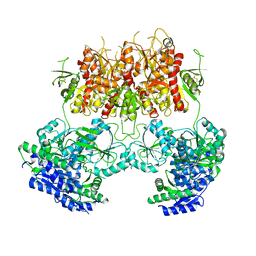 | |
7YFR
 
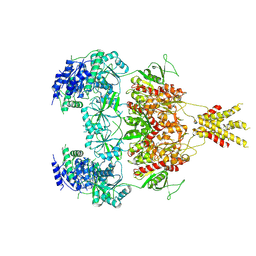 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, J.L, Zhu, S.J, Zhang, M. | | 登録日 | 2022-07-09 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFM
 
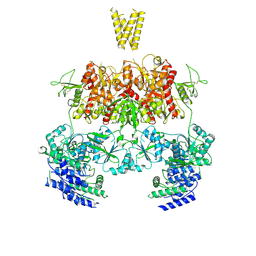 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | 分子名称: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | 著者 | Zhang, J.L, Zhu, S.J, Zhang, M. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WK2
 
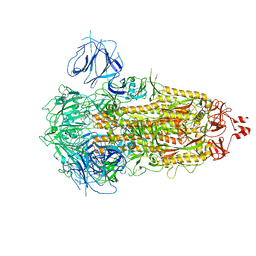 | | SARS-CoV-2 Omicron S-close | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-01-08 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WKA
 
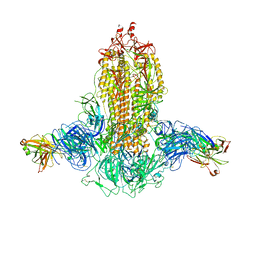 | |
7WK9
 
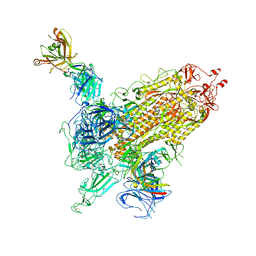 | |
7WK8
 
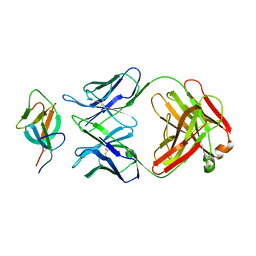 | |
7WSW
 
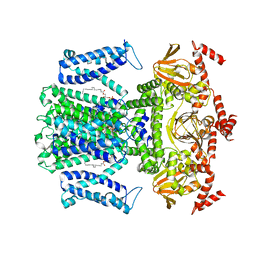 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | 分子名称: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | 著者 | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | 登録日 | 2022-02-02 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7WN2
 
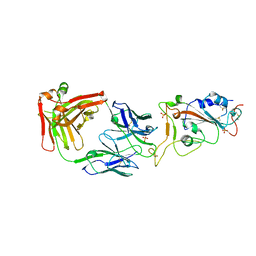 | |
7WNB
 
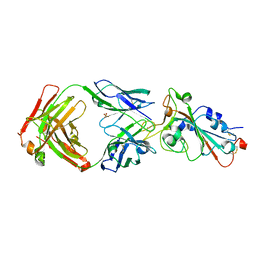 | |
4G78
 
 | |
7XUF
 
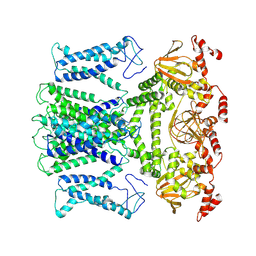 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | 分子名称: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | 著者 | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | 登録日 | 2022-05-18 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
5TO3
 
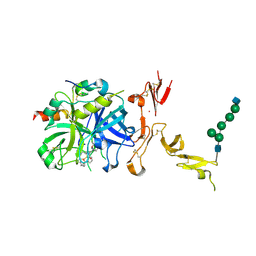 | | Crystal structure of thrombin mutant W215A/E217A fused to EGF456 of thrombomodulin via a 31-residue linker and bound to PPACK | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | 著者 | Barranco-Medina, S, Murphy, M, Pelc, L, Chen, Z, Di Cera, E, Pozzi, N. | | 登録日 | 2016-10-16 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Rational Design of Protein C Activators.
Sci Rep, 7, 2017
|
|
3WAJ
 
 | |
3WAK
 
 | |
7WV9
 
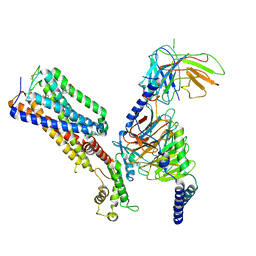 | | Allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 in complex with Gi protein | | 分子名称: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, ... | | 著者 | Xu, Z, Shao, Z. | | 登録日 | 2022-02-10 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
7XGW
 
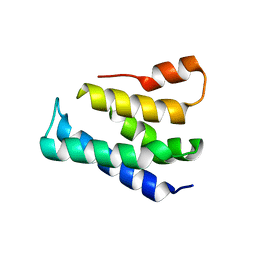 | |
7XY9
 
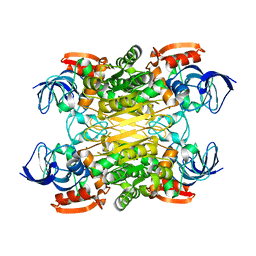 | | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | | 分子名称: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, ZINC ION | | 著者 | Chen, Q, Li, X, Yang, F, Qu, G, Sun, Z.T, Wang, Y.J. | | 登録日 | 2022-06-01 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.12 Å) | | 主引用文献 | Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions.
Nat Commun, 14, 2023
|
|
7V9P
 
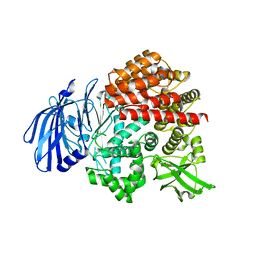 | |
7V9Q
 
 | |
7V9O
 
 | |
7V9N
 
 | | Crystal structure of the lanthipeptide zinc-metallopeptidase EryP from saccharopolyspora erythraea in closed state | | 分子名称: | ACETATE ION, Alanine aminopeptidase, CALCIUM ION, ... | | 著者 | Zhao, C, Zhao, N.L, Bao, R. | | 登録日 | 2021-08-26 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational remodeling enhances activity of lanthipeptide zinc-metallopeptidases.
Nat.Chem.Biol., 18, 2022
|
|
7WVR
 
 | |
7X1R
 
 | | Cryo-EM structure of human thioredoxin reductase bound by Au | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, Thioredoxin reductase 1, ... | | 著者 | He, Z.S, Cao, P, Cao, S.H, He, B, Jiang, H.D, Gong, Y, Gao, X.Y. | | 登録日 | 2022-02-24 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Au4 cluster inhibits human thioredoxin reductase activity via specifically binding of Au to Cys189
Nano Today, 47, 2022
|
|
