3Q0B
 
 | |
3Q0F
 
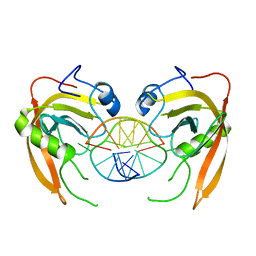 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | 分子名称: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3RAX
 
 | |
3RV1
 
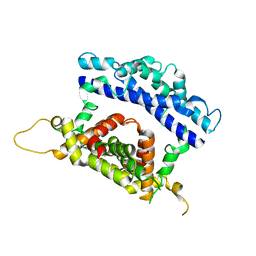 | | Crystal structure of the N-terminal and RNase III domains of K. polysporus Dcr1 E224Q mutant | | 分子名称: | K. polysporus Dcr1 | | 著者 | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | 登録日 | 2011-05-05 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.975 Å) | | 主引用文献 | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
3RW6
 
 | | Structure of nuclear RNA export factor TAP bound to CTE RNA | | 分子名称: | Nuclear RNA export factor 1, constitutive transport element(CTE)of Mason-Pfizer monkey virus RNA | | 著者 | Teplova, M, Khin, N.W, Patel, D.J, Izaurralde, E. | | 登録日 | 2011-05-07 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3OWZ
 
 | | Crystal structure of glycine riboswitch, soaked in Iridium | | 分子名称: | Domain II of glycine riboswitch, GLYCINE, IRIDIUM HEXAMMINE ION, ... | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2010-09-20 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.949 Å) | | 主引用文献 | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXB
 
 | |
3OXM
 
 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | 分子名称: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2010-09-21 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3RWV
 
 | | Crystal Structure of apo-form of Human Glycolipid Transfer Protein at 1.5 A resolution | | 分子名称: | Glycolipid transfer protein, SULFATE ION | | 著者 | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | 登録日 | 2011-05-09 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0K
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | 分子名称: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | 著者 | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | 登録日 | 2011-05-13 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3RW7
 
 | | Structure of N-terminal domain of nuclear RNA export factor TAP | | 分子名称: | Nuclear RNA export factor 1 | | 著者 | Teplova, M, Khin, N.W, Wohlbold, L, Izaurralde, E, Patel, D.J. | | 登録日 | 2011-05-07 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3S0I
 
 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | 分子名称: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | 著者 | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | 登録日 | 2011-05-13 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3RV0
 
 | | Crystal structure of K. polysporus Dcr1 without the C-terminal dsRBD | | 分子名称: | K. polysporus Dcr1, MAGNESIUM ION | | 著者 | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | 登録日 | 2011-05-05 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
6OV0
 
 | |
6PPR
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | 分子名称: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | 著者 | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | 登録日 | 2019-07-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PPJ
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | | 分子名称: | ATP-dependent DNA helicase (UvrD/REP), IRON/SULFUR CLUSTER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | 登録日 | 2019-07-07 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6D92
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-A non-canonical pair at position 3 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*AP*C)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-27 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D8F
 
 | | RsAgo Ternary Complex with Guide RNA and Target DNA Containing T-T Bulge Within the Seed Segment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*TP*TP*TP*AP*AP*C)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-26 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D8P
 
 | | Ternary RsAgo Complex Containing Guide RNA Paired with Target DNA | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CACODYLATE ION, ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-26 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D9L
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing G-A Non-canonical Pair | | 分子名称: | DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*AP*GP*CP*AP*GP*TP*AP*AP*C)-3'), MAGNESIUM ION, RNA (5'-R(P*UP*UP*AP*CP*UP*GP*CP*GP*CP*AP*GP*GP*UP*GP*AP*CP*GP*A)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-30 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D95
 
 | | Ternary RsAgo Complex with Guide RNA Paired and Target DNA containing A8-A8' Non-Canonical Pair | | 分子名称: | DNA 24-Mer, MAGNESIUM ION, RNA (5'-R(P*UP*UP*AP*CP*UP*GP*CP*AP*CP*AP*GP*GP*UP*GP*AP*CP*GP*A)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-27 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6PPU
 
 | | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | | 分子名称: | ATP-dependent DNA helicase (UvrD/REP), DNA (29-MER), IRON/SULFUR CLUSTER, ... | | 著者 | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | 登録日 | 2019-07-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
107D
 
 | | SOLUTION STRUCTURE OF THE COVALENT DUOCARMYCIN A-DNA DUPLEX COMPLEX | | 分子名称: | 4-HYDROXY-2,8-DIMETHYL-1-OXO-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-1,2,3,6,7,8-HEXAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*CP*CP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*GP*G)-3') | | 著者 | Lin, C.H, Patel, D.J. | | 登録日 | 1995-01-17 | | 公開日 | 1995-05-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the covalent duocarmycin A-DNA duplex complex.
J.Mol.Biol., 248, 1995
|
|
143D
 
 | |
6D9K
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-G Non-canonical Pair | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*GP*GP*CP*AP*GP*TP*AP*AP*C)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-30 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
