8FA6
 
 | | Crystal structure of Ky15.10 Fab in complex with circumsporozoite protein DND peptide | | 分子名称: | 1,2-ETHANEDIOL, Circumsporozoite protein DND peptide, Ky15.10 Antibody, ... | | 著者 | Thai, E, Prieto, K, Julien, J.P. | | 登録日 | 2022-11-25 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FA7
 
 | |
8FB5
 
 | | Crystal structure of Ky15.11-S100IK Fab in complex with circumsporozoite protein KQPA peptide | | 分子名称: | Circumsporozoite protein KQPA peptide, Ky15.11-SK Antibody, heavy chain, ... | | 著者 | Kang, R.W, Thai, E, Julien, J.P. | | 登録日 | 2022-11-29 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FB6
 
 | | Crystal structure of Ky15.10 Fab in complex with circumsporozoite protein KQPA peptide | | 分子名称: | 1,2-ETHANEDIOL, Circumsporozoite protein KQPA peptide, Ky15.10 Antibody, ... | | 著者 | Prieto, K, Thai, E, Julien, J.P. | | 登録日 | 2022-11-29 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FB7
 
 | | Crystal structure of Ky15.10 Fab in complex with circumsporozoite protein NPDP peptide | | 分子名称: | 1,2-ETHANEDIOL, Circumsporozoite protein NPDP peptide, Ky15.10 Antibody, ... | | 著者 | Prieto, K, Thai, E, Julien, J.P. | | 登録日 | 2022-11-29 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FBA
 
 | |
8FA9
 
 | | Crystal structure of Ky15.5 Fab in complex with circumsporozoite protein NPDP peptide | | 分子名称: | Circumsporozoite protein NPDP peptide, Ky15.5 Antibody, heavy chain, ... | | 著者 | Thai, E, Prieto, K, Julien, J.P. | | 登録日 | 2022-11-25 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FB8
 
 | | Crystal structure of Ky15.10-Y100EK Fab in complex with circumsporozoite protein KQPA peptide | | 分子名称: | Circumsporozoite protein KQPA peptide, GLYCEROL, Ky15.10-YK Antibody, ... | | 著者 | Kang, R.W, Thai, E, Julien, J.P. | | 登録日 | 2022-11-29 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FDD
 
 | |
8FA8
 
 | |
8FAS
 
 | | Crystal structure of Ky230 Fab in complex with circumsporozoite protein NANP5 peptide | | 分子名称: | 1,2-ETHANEDIOL, Circumsporozoite protein NANP5 peptide, Ky230 Antibody, ... | | 著者 | Kassardjian, A, Thai, E, Julien, J.P. | | 登録日 | 2022-11-28 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FDC
 
 | |
6PUA
 
 | | The 2.0 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-07-18 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
6MDD
 
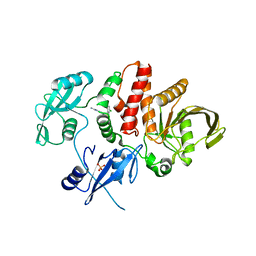 | |
8C9J
 
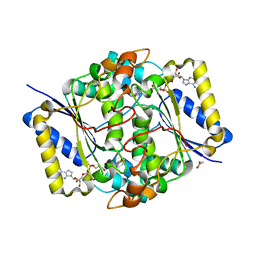 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | 分子名称: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | 著者 | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | 登録日 | 2023-01-23 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
6EG3
 
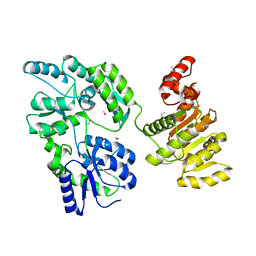 | | Crystal structure of human BRM in complex with compound 15 | | 分子名称: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | 著者 | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | 登録日 | 2018-08-17 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
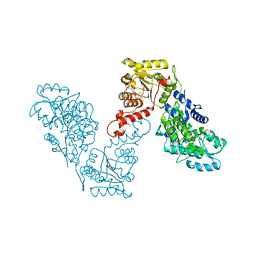 | | Crystal structure of human BRM in complex with compound 16 | | 分子名称: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | 著者 | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | 登録日 | 2018-08-17 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
3USB
 
 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | 分子名称: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-11-23 | | 公開日 | 2011-12-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
8FG0
 
 | |
6CDX
 
 | | High-resolution crystal structure of fluoropropylated cystine knot, binding to alpha-5 beta-6 integrin | | 分子名称: | cystine knot (fluoropropylated) | | 著者 | Kimura, R, Nix, J, Bongura, C, Chakraborti, S, Gambhir, S, Filipp, F.V. | | 登録日 | 2018-02-09 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Evaluation of integrin alpha v beta6cystine knot PET tracers to detect cancer and idiopathic pulmonary fibrosis.
Nat Commun, 10, 2019
|
|
7EZ0
 
 | | Apo L-21 ScaI Tetrahymena ribozyme | | 分子名称: | Apo L-21 ScaI Tetrahymena ribozyme, MAGNESIUM ION | | 著者 | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | 登録日 | 2021-06-01 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
7EZ2
 
 | | Holo L-16 ScaI Tetrahymena ribozyme | | 分子名称: | Holo L-16 ScaI Tetrahymena ribozyme, Holo L-16 ScaI Tetrahymena ribozyme S1, Holo L-16 ScaI Tetrahymena ribozyme S2, ... | | 著者 | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | 登録日 | 2021-06-01 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
7DVK
 
 | |
7DVI
 
 | |
7BBJ
 
 | |
