6KIY
 
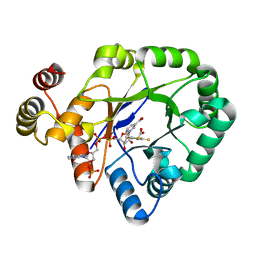 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | 著者 | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KY6
 
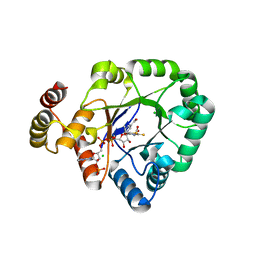 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | 分子名称: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | 登録日 | 2019-09-16 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
5ZWZ
 
 | |
5ZWX
 
 | |
6BOJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN5004 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-{[2-(3-chlorophenyl)-6-ethylpyrimidin-4-yl]methyl}phenyl)acetamide, CHLORIDE ION, ... | | 著者 | Fox III, D, Fairman, J.W, Gurney, M.E. | | 登録日 | 2017-11-20 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Memory enhancing effects of BPN14770, an allosteric inhibitor of phosphodiesterase-4D, in wild-type and humanized mice.
Neuropsychopharmacology, 43, 2018
|
|
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | 分子名称: | Dipeptide permease D | | 著者 | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | 登録日 | 2014-04-21 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
7Y7M
 
 | |
4LGH
 
 | |
4LGG
 
 | |
5W93
 
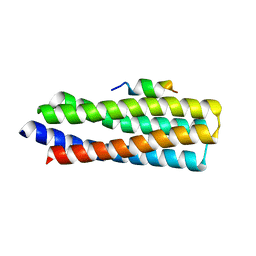 | | p130Cas complex with paxillin LD1 | | 分子名称: | Breast cancer anti-estrogen resistance protein 1, Paxillin | | 著者 | Miller, D.J, Zheng, J.J. | | 登録日 | 2017-06-22 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural and functional insights into the interaction between the Cas family scaffolding protein p130Cas and the focal adhesion-associated protein paxillin.
J. Biol. Chem., 292, 2017
|
|
7YMS
 
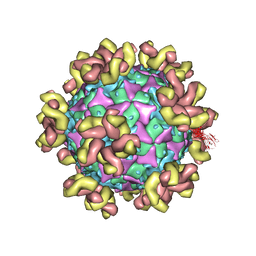 | |
7YRH
 
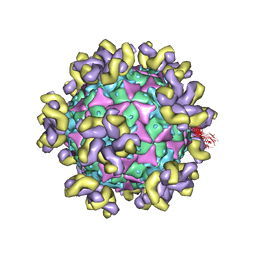 | |
7YRF
 
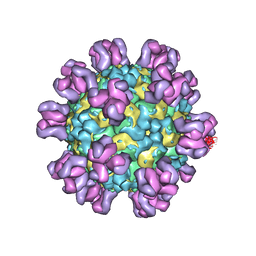 | |
7YV7
 
 | |
7YV2
 
 | |
3D0R
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group P2(1) | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Protein CalG3 | | 著者 | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | 登録日 | 2008-05-02 | | 公開日 | 2008-06-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
7RX6
 
 | |
7RX7
 
 | |
7RX8
 
 | |
3D0Q
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group I222 | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Protein CalG3 | | 著者 | Bitto, E, Singh, S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | 登録日 | 2008-05-02 | | 公開日 | 2008-06-24 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
7DD8
 
 | |
7DK5
 
 | |
7DK6
 
 | |
7DCC
 
 | |
7DD2
 
 | |
