4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | 分子名称: | CALCIUM ION, QDE-2-interacting protein | | 著者 | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | 登録日 | 2014-09-03 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5H1M
 
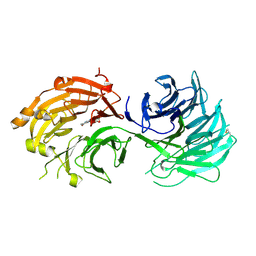 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with M7G | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Gem-associated protein 5 | | 著者 | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | 登録日 | 2016-10-10 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.492 Å) | | 主引用文献 | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1K
 
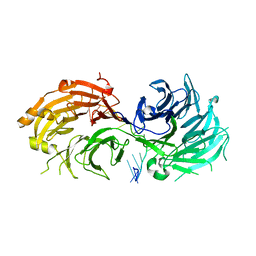 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 13-nt U4 snRNA fragment | | 分子名称: | Gem-associated protein 5, U4 snRNA (5'-R(*GP*CP*AP*AP*UP*UP*UP*UP*UP*GP*AP*CP*A)-3') | | 著者 | Wang, Y, Jin, W, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | 登録日 | 2016-10-10 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5JWY
 
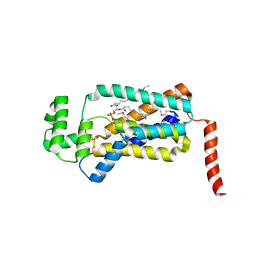 | | Structure of lipid phosphate phosphatase PgpB complex with PE | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Phosphatidylglycerophosphatase B | | 著者 | Tong, S, Wang, M, Zheng, L. | | 登録日 | 2016-05-12 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Insight into Substrate Selection and Catalysis of Lipid Phosphate Phosphatase PgpB in the Cell Membrane.
J.Biol.Chem., 291, 2016
|
|
5YEL
 
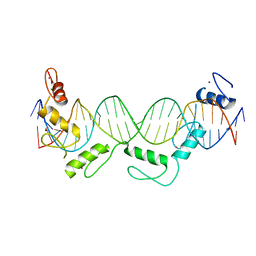 | | Crystal structure of CTCF ZFs6-11-gb7CSE | | 分子名称: | DNA (26-MER), Transcriptional repressor CTCF, ZINC ION | | 著者 | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | 登録日 | 2017-09-18 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
8KDB
 
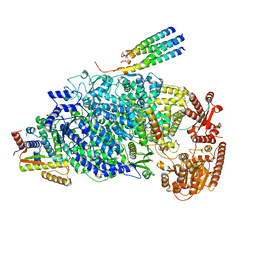 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form | | 分子名称: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | 著者 | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | 登録日 | 2023-08-09 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
8KDC
 
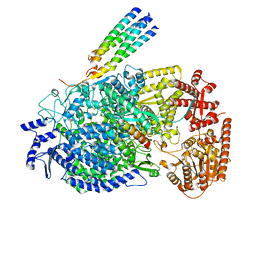 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form | | 分子名称: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | 著者 | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | 登録日 | 2023-08-09 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
5YEH
 
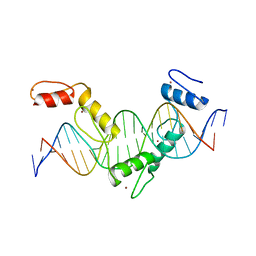 | | Crystal structure of CTCF ZFs4-8-eCBS | | 分子名称: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | 登録日 | 2017-09-17 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.328 Å) | | 主引用文献 | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5YEF
 
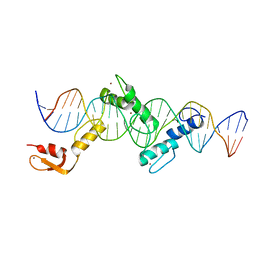 | | Crystal structure of CTCF ZFs2-8-Hs5-1aE | | 分子名称: | DNA (27-MER), Transcriptional repressor CTCF, ZINC ION | | 著者 | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | 登録日 | 2017-09-17 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.807 Å) | | 主引用文献 | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5H1J
 
 | | Crystal structure of WD40 repeat domains of Gemin5 | | 分子名称: | Gem-associated protein 5 | | 著者 | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | 登録日 | 2016-10-10 | | 公開日 | 2016-11-23 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
3CFP
 
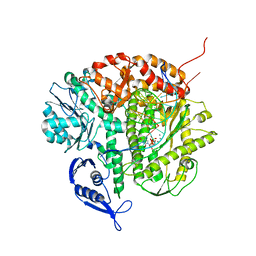 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 1 | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), ... | | 著者 | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | 登録日 | 2008-03-04 | | 公開日 | 2009-03-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
4ATD
 
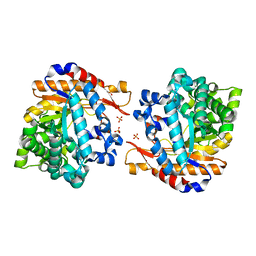 | | Crystal structure of native Raucaffricine glucosidase | | 分子名称: | RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | 著者 | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | 登録日 | 2012-05-05 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | High Speed X-Ray Analysis of Plant Enzymes at Room Temperature
Phytochemistry, 91, 2013
|
|
4ATL
 
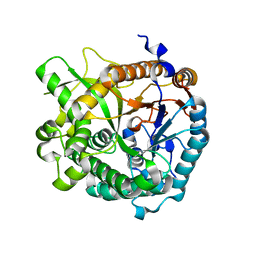 | | Crystal structure of Raucaffricine glucosidase in complex with Glucose | | 分子名称: | RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, beta-D-glucopyranose | | 著者 | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | 登録日 | 2012-05-08 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | High Speed X-Ray Analysis of Plant Enzymes at Room Temperature
Phytochemistry, 91, 2013
|
|
3CFO
 
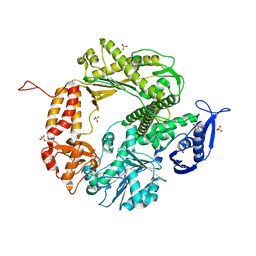 | | Triple Mutant APO structure | | 分子名称: | DNA polymerase, GUANOSINE, SULFATE ION | | 著者 | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | 登録日 | 2008-03-04 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
5LIS
 
 | |
5LIN
 
 | |
4U7U
 
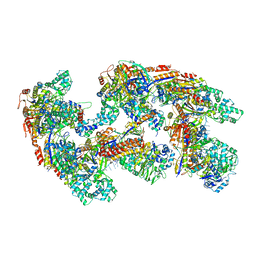 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | 分子名称: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | 著者 | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | 登録日 | 2014-07-31 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.003 Å) | | 主引用文献 | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
5LIO
 
 | |
5H1L
 
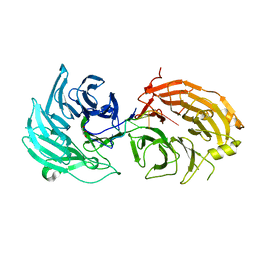 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 7-nt U4 snRNA fragment | | 分子名称: | GLYCEROL, Gem-associated protein 5, U4 snRNA (5'-R(*AP*UP*UP*UP*UP*UP*G)-3') | | 著者 | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | 登録日 | 2016-10-10 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
3ZJ6
 
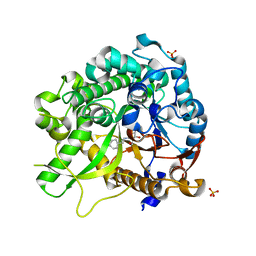 | | Crystal of Raucaffricine Glucosidase in complex with inhibitor | | 分子名称: | (1R,2S,3S,4R,5R)-4-(cyclohexylmethylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3CFR
 
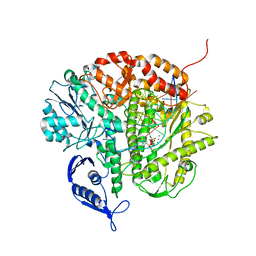 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 2 | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*(DOC))-3'), ... | | 著者 | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | 登録日 | 2008-03-04 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
6LI4
 
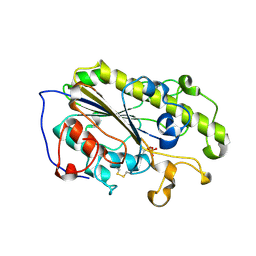 | | Crystal structure of MCR-1-S | | 分子名称: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | 著者 | Zhang, Q, Wang, M, Sun, H. | | 登録日 | 2019-12-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI5
 
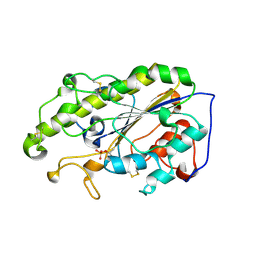 | | Crystal structure of apo-MCR-1-S | | 分子名称: | Probable phosphatidylethanolamine transferase Mcr-1 | | 著者 | Zhang, Q, Wang, M, Sun, H. | | 登録日 | 2019-12-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
5YEG
 
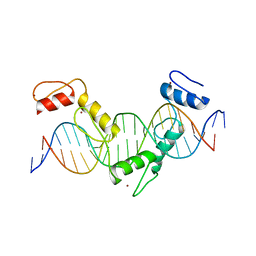 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | 分子名称: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Yin, M, Wang, J, Wang, M, Li, X. | | 登録日 | 2017-09-17 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
8XJ3
 
 | |
