3IQS
 
 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | 著者 | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | 登録日 | 2009-08-20 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
1K6O
 
 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | 分子名称: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | 著者 | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | 登録日 | 2001-10-16 | | 公開日 | 2002-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
2KEI
 
 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | 分子名称: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | 著者 | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | 登録日 | 2009-01-30 | | 公開日 | 2009-05-19 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEK
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | 分子名称: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | 著者 | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | 登録日 | 2009-01-30 | | 公開日 | 2009-05-19 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEJ
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | 分子名称: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | 著者 | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | 登録日 | 2009-01-30 | | 公開日 | 2009-05-19 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
1O7A
 
 | | Human beta-Hexosaminidase B | | 分子名称: | 1,2-ETHANEDIOL, 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Maier, T, Strater, N, Schuette, C, Klingenstein, R, Sandhoff, K, Saenger, W. | | 登録日 | 2002-10-29 | | 公開日 | 2003-10-23 | | 最終更新日 | 2020-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The X-Ray Crystal Structure of Human Beta-Hexosaminidase B Provides New Insights Into Sandhoff Disease
J.Mol.Biol., 328, 2003
|
|
2KN7
 
 | | Structure of the XPF-single strand DNA complex | | 分子名称: | DNA (5'-D(*CP*AP*GP*TP*GP*GP*CP*TP*GP*A)-3'), DNA repair endonuclease XPF | | 著者 | Das, D, Folkers, G.E, van Dijk, M, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | 登録日 | 2009-08-16 | | 公開日 | 2010-08-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the XPF-ssDNA complex underscores the distinct roles of the XPF and ERCC1 helix- hairpin-helix domains in ss/ds DNA recognition
Structure, 20, 2012
|
|
1M1D
 
 | | TETRAHYMENA GCN5 WITH BOUND BISUBSTRATE ANALOG INHIBITOR | | 分子名称: | HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | 著者 | Poux, A.N, Cebrat, M, Kim, C.M, Cole, P.A, Marmorstein, R. | | 登録日 | 2002-06-18 | | 公開日 | 2002-10-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3LBX
 
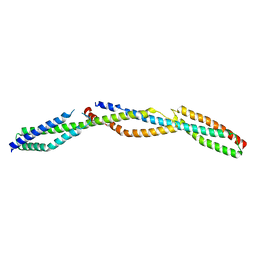 | | Crystal Structure of the Erythrocyte Spectrin Tetramerization Domain Complex | | 分子名称: | Spectrin alpha chain, erythrocyte, Spectrin beta chain | | 著者 | Ipsaro, J.J, Harper, S.L, Messick, T.E, Marmorstein, R, Mondragon, A, Speicher, D.W. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure and functional interpretation of the erythrocyte spectrin tetramerization domain complex.
Blood, 115, 2010
|
|
1MJA
 
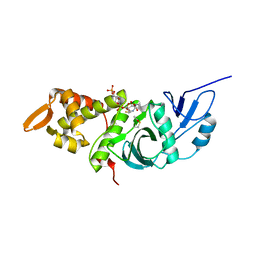 | | Crystal structure of yeast Esa1 histone acetyltransferase domain complexed with acetyl coenzyme A | | 分子名称: | COENZYME A, Esa1 protein | | 著者 | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | 登録日 | 2002-08-27 | | 公開日 | 2002-10-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1MJ9
 
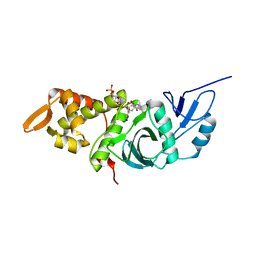 | | Crystal structure of yeast Esa1(C304S) mutant complexed with Coenzyme A | | 分子名称: | COENZYME A, ESA1 PROTEIN, SODIUM ION | | 著者 | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | 登録日 | 2002-08-27 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
2JPD
 
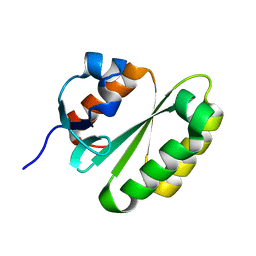 | | Solution structure of the ERCC1 central domain | | 分子名称: | DNA excision repair protein ERCC-1 | | 著者 | Tripsianes, K, Folkers, G, Zheng, C, Das, D, Grinstead, J.S, Kaptein, R, Boelens, R. | | 登録日 | 2007-05-06 | | 公開日 | 2007-09-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization
Nucleic Acids Res., 35, 2007
|
|
1MJB
 
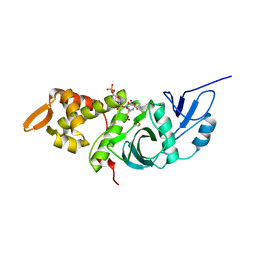 | | Crystal structure of yeast Esa1 histone acetyltransferase E338Q mutant complexed with acetyl coenzyme A | | 分子名称: | ACETYL COENZYME *A, Esa1 protein | | 著者 | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | 登録日 | 2002-08-27 | | 公開日 | 2002-10-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
3QM0
 
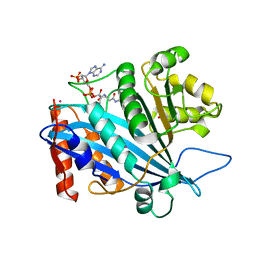 | | Crystal structure of RTT109-AC-CoA complex | | 分子名称: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | 著者 | Tang, Y, Marmorstein, R. | | 登録日 | 2011-02-03 | | 公開日 | 2011-02-16 | | 最終更新日 | 2017-08-02 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1SQT
 
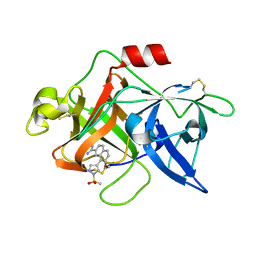 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | 分子名称: | 7-METHOXY-8-[1-(METHYLSULFONYL)-1H-PYRAZOL-4-YL]NAPHTHALENE-2-CARBOXIMIDAMIDE, Urokinase-type plasminogen activator | | 著者 | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhang, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | 登録日 | 2004-03-19 | | 公開日 | 2004-04-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S5P
 
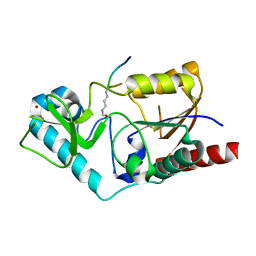 | | Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Eschericia coli. | | 分子名称: | HISTONE H4 (RESIDUES 12-19), NAD-dependent deacetylase, ZINC ION | | 著者 | Zhao, K, Chai, X, Marmorstein, R. | | 登録日 | 2004-01-21 | | 公開日 | 2004-03-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structure and Substrate Binding Properties of cobB, a Sir2 Homolog Protein Deacetylase from Eschericia coli.
J.Mol.Biol., 337, 2004
|
|
3RQ3
 
 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form | | 分子名称: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | 著者 | Ramagopal, U.A, Rubinstein, R, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | 登録日 | 2011-04-27 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form
To be published
|
|
1CS3
 
 | |
1BC7
 
 | | SERUM RESPONSE FACTOR ACCESSORY PROTEIN 1A (SAP-1)/DNA COMPLEX | | 分子名称: | DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*AP*GP*GP*AP*TP*GP*TP*G)-3'), PROTEIN (ETS-DOMAIN PROTEIN) | | 著者 | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | 登録日 | 1998-05-05 | | 公開日 | 1999-01-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
1B06
 
 | | SUPEROXIDE DISMUTASE FROM SULFOLOBUS ACIDOCALDARIUS | | 分子名称: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | 著者 | Knapp, S, Kardinahl, S, Niklas, H, Tibbelin, G, Schafer, G, Ladenstein, R. | | 登録日 | 1998-11-16 | | 公開日 | 1999-11-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Refined crystal structure of a superoxide dismutase from the hyperthermophilic archaeon Sulfolobus acidocaldarius at 2.2 A resolution.
J.Mol.Biol., 285, 1999
|
|
1B3B
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT N97D, G376K | | 分子名称: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | 著者 | Knapp, S, Lebbink, J.H.G, Van Der Oost, J, Devos, W.M, Rice, D, Ladenstein, R. | | 登録日 | 1998-12-07 | | 公開日 | 1999-12-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. I. Introduction of a six-residue ion-pair network in the hinge region.
J.Mol.Biol., 280, 1998
|
|
1BC8
 
 | | STRUCTURES OF SAP-1 BOUND TO DNA SEQUENCES FROM THE E74 AND C-FOS PROMOTERS PROVIDE INSIGHTS INTO HOW ETS PROTEINS DISCRIMINATE BETWEEN RELATED DNA TARGETS | | 分子名称: | DNA (5'-D(*AP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), PROTEIN (SAP-1 ETS DOMAIN), ... | | 著者 | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | 登録日 | 1998-05-05 | | 公開日 | 1998-12-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
1B26
 
 | | GLUTAMATE DEHYDROGENASE | | 分子名称: | GLUTAMATE DEHYDROGENASE | | 著者 | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | 登録日 | 1998-12-04 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
3TFY
 
 | |
1DUX
 
 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | 分子名称: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | 著者 | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | 登録日 | 2000-01-19 | | 公開日 | 2000-04-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
