8HBT
 
 | | AmAT7-3 mutant A310G | | 分子名称: | AmAT7-3-A310G, Astragaloside IV | | 著者 | Wang, L.L. | | 登録日 | 2022-10-31 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
8H8I
 
 | | Triterpenoid saponin acetyltransferase, AmAT7-3 | | 分子名称: | AmAT7-3, Astragaloside IV | | 著者 | Wang, L.L. | | 登録日 | 2022-10-22 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
6JFX
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltopentaose | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | 登録日 | 2019-02-12 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.981 Å) | | 主引用文献 | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JHF
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | 登録日 | 2019-02-18 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JEQ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with beta-cyclodextrin | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | 著者 | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | 登録日 | 2019-02-07 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6A7B
 
 | | AKR1C3 complexed with new inhibitor with novel scaffold | | 分子名称: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | 著者 | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | 登録日 | 2018-07-02 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
7FAR
 
 | | Crystal structure of PDE5A in complex with inhibitor L12 | | 分子名称: | 5-[bis(fluoranyl)methoxy]-2-[(4-chlorophenyl)methyl]-10-(3-methoxypropyl)-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, ZINC ION, ... | | 著者 | Wu, D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.40006471 Å) | | 主引用文献 | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
7FAQ
 
 | | Crystal structure of PDE5A in complex with inhibitor L1 | | 分子名称: | 2-[(4-chlorophenyl)methyl]-5-methoxy-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Wu, D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.200136 Å) | | 主引用文献 | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
8WIU
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | 分子名称: | 7-[5-[1-(cyclopropylmethyl)-3,5-dimethyl-pyrazol-4-yl]pyridin-3-yl]-1~{H}-imidazo[4,5-b]pyridine, Isoform C of Bromodomain-containing protein 4 | | 著者 | Cao, D, Zhiyan, D, Xiong, B. | | 登録日 | 2023-09-25 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Discovery of a brain-permeable bromodomain and extra terminal domain (BET) inhibitor with selectivity for BD1 for the treatment of multiple sclerosis.
Eur.J.Med.Chem., 265, 2023
|
|
3RN2
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Immune Receptors | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*TP*CP*TP*TP*TP*GP*AP*TP*GP*G)-3'), Interferon-inducible protein AIM2 | | 著者 | Jin, T.C, Xiao, T. | | 登録日 | 2011-04-21 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RLN
 
 | |
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | 分子名称: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | 登録日 | 2018-10-12 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
3RLO
 
 | |
3RN5
 
 | | Structural basis of cytosolic DNA recognition by innate immune receptors | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*AP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*TP*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*G)-3'), ... | | 著者 | Jin, T.C, Xiao, T. | | 登録日 | 2011-04-21 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RNU
 
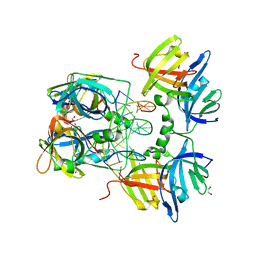 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Jin, T.C, Xiao, T. | | 登録日 | 2011-04-22 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
5YLH
 
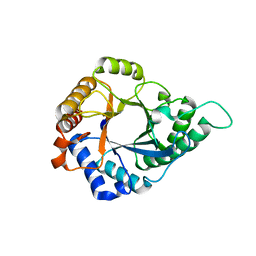 | | Structure of GH113 beta-1,4-mannanase | | 分子名称: | beta-1,4-mannanase | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-10-17 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YLL
 
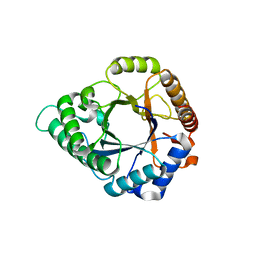 | | Structure of GH113 beta-1,4-mannanase complex with M6. | | 分子名称: | beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-10-17 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YLI
 
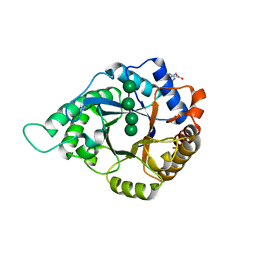 | | Complex structure of GH113 beta-1,4-mannanase | | 分子名称: | beta-1,4-mannanas, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-10-17 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YLK
 
 | | Complex structure of GH 113 family beta-1,4-mannanase with mannobiose | | 分子名称: | beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-10-17 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase fromn Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5Z4T
 
 | | Complex structure - AxMan113A-M3 | | 分子名称: | beta-1,4-mannanas, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2018-01-13 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase fromAmphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
6AIC
 
 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | 著者 | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | 登録日 | 2018-08-22 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7RMR
 
 | |
7RMQ
 
 | | Crystal structure of cycloviolacin O2 | | 分子名称: | Cycloviolacin O2, D-[I11L]cycloviolacin O2, FORMIC ACID | | 著者 | Huang, Y.H, Du, Q. | | 登録日 | 2021-07-28 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMS
 
 | |
6KFW
 
 | |
