7ZS5
 
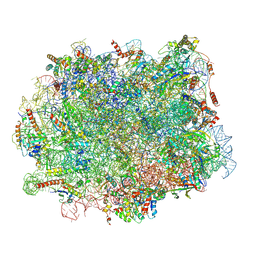 | | Structure of 60S ribosomal subunit from S. cerevisiae with eIF6 and tRNA | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-06 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZRS
 
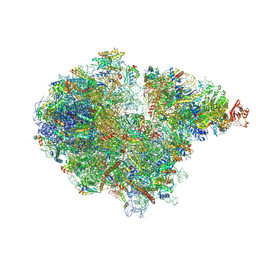 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C2) - composite map | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-05 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUX
 
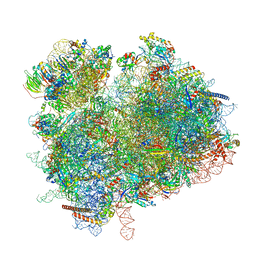 | | Collided ribosome in a disome unit from S. cerevisiae | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUW
 
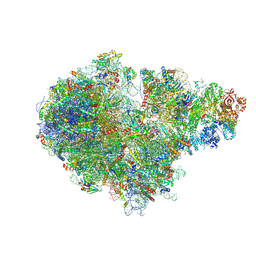 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZJ4
 
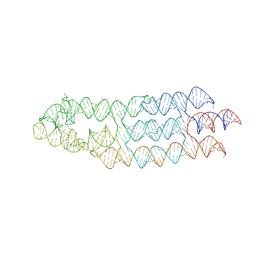 | | Ligand bound state of a brocolli-pepper aptamer FRET tile | | 分子名称: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, POTASSIUM ION, ... | | 著者 | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | 登録日 | 2022-04-08 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
8A58
 
 | |
8A9P
 
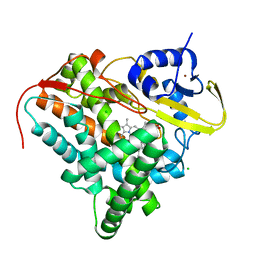 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment | | 分子名称: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Snee, M, Katariya, M, Levy, C, Leys, D. | | 登録日 | 2022-06-29 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment
To Be Published
|
|
7ZJ5
 
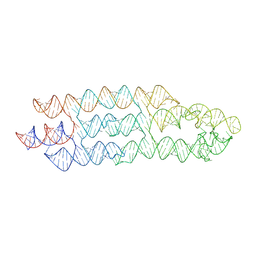 | | Unbound state of a brocolli-pepper aptamer FRET tile. | | 分子名称: | POTASSIUM ION, brocolli-pepper aptamer | | 著者 | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | 登録日 | 2022-04-08 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.55 Å) | | 主引用文献 | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
7ZM5
 
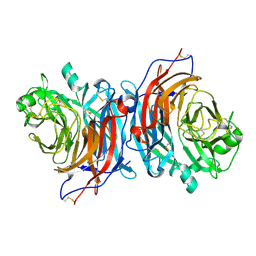 | | Structure of Mossman virus receptor binding protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | 著者 | Stelfox, A.J, Bowden, T.A, Rissanen, I, Harlos, K. | | 登録日 | 2022-04-19 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Mbio, 14, 2023
|
|
7ZJV
 
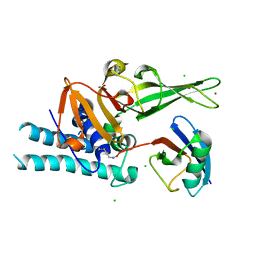 | |
6YLV
 
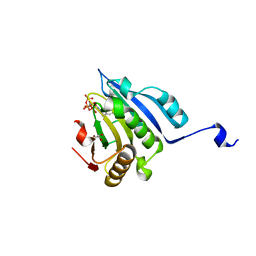 | | Translation initiation factor 4E in complex with 4-Cl-Bn7GpppG mRNA 5' cap analog | | 分子名称: | 4-Cl-Bn7GpppG mRNA 5' cap analog, Eukaryotic translation initiation factor 4E, GLYCEROL | | 著者 | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | 登録日 | 2020-04-07 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.66005659 Å) | | 主引用文献 | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6ZEZ
 
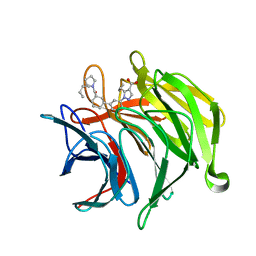 | |
6ZF5
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | 分子名称: | 1-[3-[(4-butylphenyl)sulfonyl-(2-hydroxy-2-oxoethyl)amino]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | 著者 | Narayanan, D, Bach, A, Gajhede, M. | | 登録日 | 2020-06-16 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZEX
 
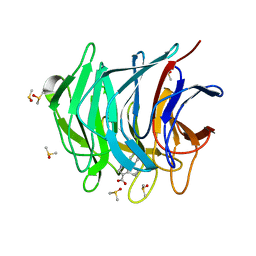 | | Keap1 kelch domain bound to a small molecule fragment | | 分子名称: | 5-cyclopropyl-1-phenyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | 著者 | Narayanan, D, Bach, A, Gajhede, M. | | 登録日 | 2020-06-16 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6Z2H
 
 | |
7A9I
 
 | |
7A9J
 
 | |
7ABB
 
 | |
7BJ3
 
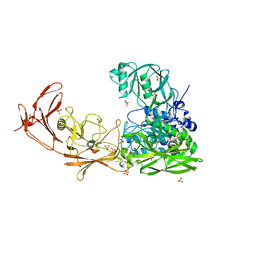 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | 著者 | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | 登録日 | 2021-01-13 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
7A1A
 
 | |
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | 著者 | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7AYG
 
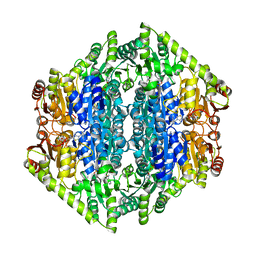 | | oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | 著者 | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | 登録日 | 2020-11-12 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7B2E
 
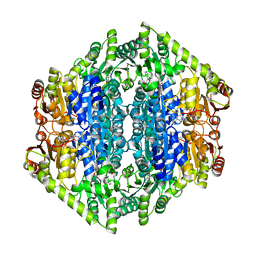 | | quadruple mutant of oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | 著者 | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | 登録日 | 2020-11-26 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7DZ5
 
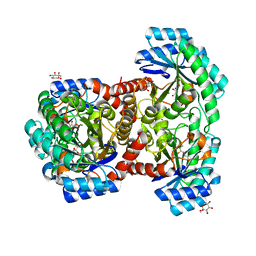 | | Crystal structures of D-allulose 3-epimerase with D-sorbose from Sinorhizobium fredii | | 分子名称: | D-sorbose, D-tagatose 3-epimerase, MAGNESIUM ION, ... | | 著者 | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | 登録日 | 2021-01-23 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7T69
 
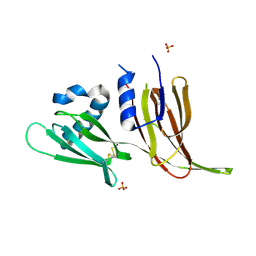 | | Crystal structure of Avr3 (SIX1) from Fusarium oxysporum f. sp. lycopersici | | 分子名称: | Avr3 (SIX1), Secreted in xylem 1, SULFATE ION | | 著者 | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | 登録日 | 2021-12-13 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
