6NJN
 
 | |
6NJM
 
 | |
5YJ3
 
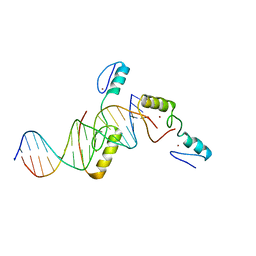 | | Crystal structure of TZAP and telomeric DNA complex | | 分子名称: | DNA (5'-D(*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), Telomere zinc finger-associated protein, ... | | 著者 | Li, F, Zhao, Y. | | 登録日 | 2017-10-07 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.845 Å) | | 主引用文献 | The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA.
Cell Res., 28, 2018
|
|
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | 分子名称: | Dipeptide permease D | | 著者 | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | 登録日 | 2014-04-21 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
7E5X
 
 | |
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp6/7 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp14/15 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VAH
 
 | |
7EKI
 
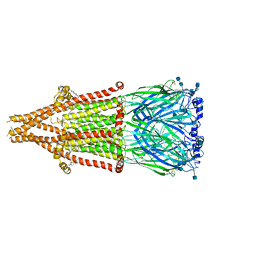 | | human alpha 7 nicotinic acetylcholine receptor in apo-form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Liu, S, Zhao, Y, Sun, D, Tian, C. | | 登録日 | 2021-04-05 | | 公開日 | 2021-05-19 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKP
 
 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | 著者 | Liu, S, Zhao, Y, Sun, D, Tian, C. | | 登録日 | 2021-04-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKT
 
 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 and PNU-120596 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | 著者 | Liu, S, Zhao, Y, Sun, D, Tian, C. | | 登録日 | 2021-04-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7DHP
 
 | |
7XR4
 
 | |
7XR6
 
 | | Structure of human excitatory amino acid transporter 2 (EAAT2) in complex with WAY-213613 | | 分子名称: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Zhao, Y, Zhang, Z. | | 登録日 | 2022-05-09 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of ligand binding modes of human EAAT2.
Nat Commun, 13, 2022
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | 分子名称: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | 著者 | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | 登録日 | 2014-08-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
6R8R
 
 | | Structure of the Wnt deacylase Notum in complex with isoquinoline 45 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Vecchia, L, Zhao, Y, Ruza, R.R, Jones, E.Y. | | 登録日 | 2019-04-02 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
1F0J
 
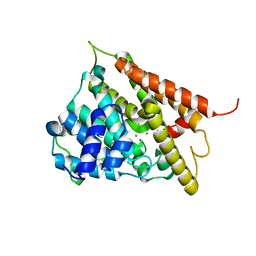 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 4B2B | | 分子名称: | ARSENIC, MAGNESIUM ION, PHOSPHODIESTERASE 4B, ... | | 著者 | Xu, R.X, Hassell, A.M, Vanderwall, D, Lambert, M.H, Holmes, W.D, Luther, M.A, Rocque, W.J, Milburn, M.V, Zhao, Y, Ke, H, Nolte, R.T. | | 登録日 | 2000-05-16 | | 公開日 | 2000-07-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity.
Science, 288, 2000
|
|
8WFJ
 
 | |
8WFK
 
 | |
8WFL
 
 | |
8WFI
 
 | |
