7X8G
 
 | |
7X8B
 
 | |
7X8F
 
 | |
7JKZ
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with YF3-126 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | 登録日 | 2020-07-29 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKY
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-126 | | 分子名称: | Bromodomain-containing protein 4, N-(1-[1,1-di(pyridin-2-yl)ethyl]-6-{1-methyl-6-oxo-5-[(piperidin-4-yl)amino]-1,6-dihydropyridin-3-yl}-1H-indol-4-yl)ethanesulfonamide, SODIUM ION | | 著者 | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | 登録日 | 2020-07-29 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKW
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with ZN1-99 | | 分子名称: | Bromodomain-containing protein 4, N-(6-{5-[(azetidin-3-yl)amino]-1-methyl-6-oxo-1,6-dihydropyridin-3-yl}-1-[1,1-di(pyridin-2-yl)ethyl]-1H-indol-4-yl)ethanesulfonamide | | 著者 | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | 登録日 | 2020-07-29 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
5Y27
 
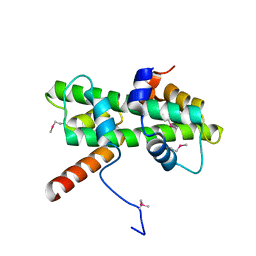 | | Crystal structure of Se-Met Dpb4-Dpb3 | | 分子名称: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | 著者 | Li, Y, Gao, F, Su, M, Zhang, F.B, Chen, Y.H. | | 登録日 | 2017-07-24 | | 公開日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8HZW
 
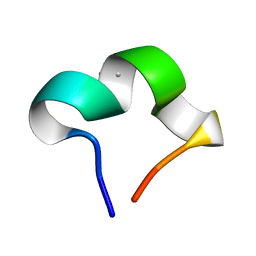 | | The NMR structure of noursinH11W peptide | | 分子名称: | noursinH11W | | 著者 | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
7WNQ
 
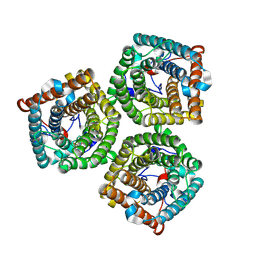 | | Cryo-EM structure of AtSLAC1 S59A mutant | | 分子名称: | Guard cell S-type anion channel SLAC1 | | 著者 | Sun, L, Liu, X, Li, Y. | | 登録日 | 2022-01-19 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structure of the Arabidopsis guard cell anion channel SLAC1 suggests activation mechanism by phosphorylation.
Nat Commun, 13, 2022
|
|
7YDX
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound RI-962 | | 分子名称: | 1-methyl-5-[2-(2-methylpropanoylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-phenylethyl]indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | 著者 | Zhang, L, Wang, Y, Li, Y, Wu, C, Luo, X, Wang, T, Lei, J, Yang, S. | | 登録日 | 2022-07-04 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.642 Å) | | 主引用文献 | Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor.
Nat Commun, 13, 2022
|
|
8JYZ
 
 | | Cryo-EM structure of RCD-1 pore from Neurospora crassa | | 分子名称: | Gasdermin-like protein rcd-1-1, Gasdermin-like protein rcd-1-2 | | 著者 | Hou, Y.J, Sun, Q, Li, Y, Ding, J. | | 登録日 | 2023-07-04 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Cleavage-independent activation of ancient eukaryotic gasdermins and structural mechanisms.
Science, 384, 2024
|
|
8I17
 
 | |
7XYT
 
 | |
7E62
 
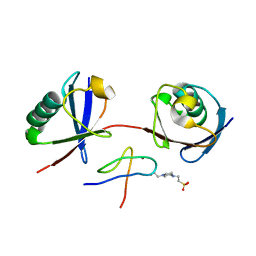 | | Mouse TAB2 NZF in complex with Lys6-linked diubiquitin | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGF-beta-activated kinase 1 and MAP3K7-binding protein 2, Ubiquitin, ... | | 著者 | Sato, Y, Li, Y, Okatsu, K, Fukai, S. | | 登録日 | 2021-02-21 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis for specific recognition of K6-linked polyubiquitin chains by the TAB2 NZF domain.
Biophys.J., 120, 2021
|
|
7YFS
 
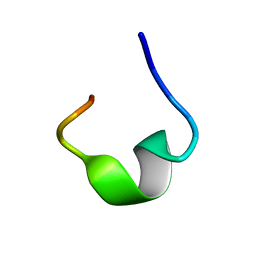 | | The NMR structure of noursin, a tricyclic ribosomal peptide containing a histidine-to-butyrine crosslink | | 分子名称: | noursin | | 著者 | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | 登録日 | 2022-07-09 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
7XYV
 
 | |
7XYW
 
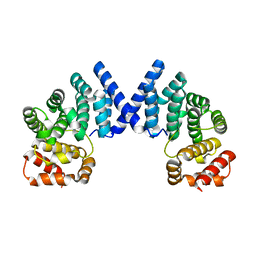 | |
7XYX
 
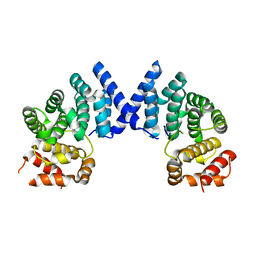 | |
7XYS
 
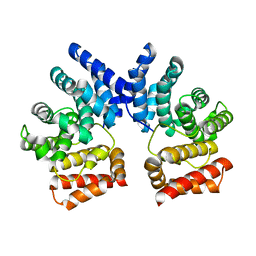 | |
7XYU
 
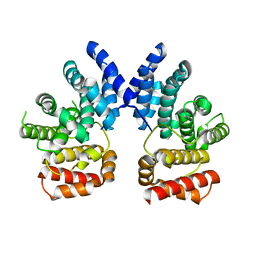 | |
7BT6
 
 | |
7BTB
 
 | |
6M62
 
 | |
6MHU
 
 | | Nucleotide-free Cryo-EM Structure of E.coli LptB2FG Transporter | | 分子名称: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Lipopolysaccharide export system ATP-binding protein LptB, Lipopolysaccharide export system permease protein LptF, ... | | 著者 | Orlando, B.J, Li, Y, Liao, M. | | 登録日 | 2018-09-18 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of lipopolysaccharide extraction by the LptB2FGC complex.
Nature, 567, 2019
|
|
6MI8
 
 | | Cryo-EM Structure of vanadate-trapped E.coli LptB2FGC | | 分子名称: | ADP ORTHOVANADATE, Lipopolysaccharide export system ATP-binding protein LptB, Lipopolysaccharide export system permease protein LptF, ... | | 著者 | Orlando, B.J, Li, Y, Liao, M. | | 登録日 | 2018-09-19 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of lipopolysaccharide extraction by the LptB2FGC complex.
Nature, 567, 2019
|
|
