7WHK
 
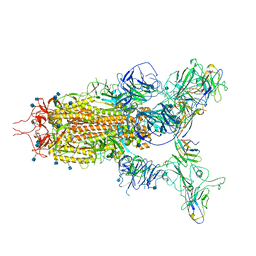 | | The state 3 complex structure of Omicron spike with Bn03 (2-up RBD, 5 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHI
 
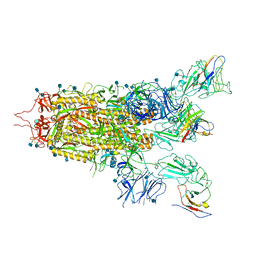 | | The state 2 complex structure of Omicron spike with Bn03 (2-up RBD, 4 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHJ
 
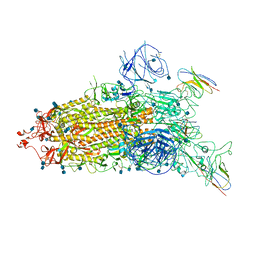 | | The state 1 complex structure of Omicron spike with Bn03 (1-up RBD, 3 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
3P96
 
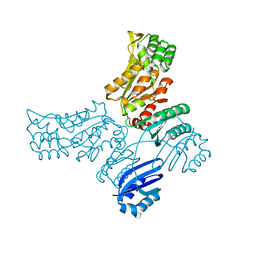 | |
7CH1
 
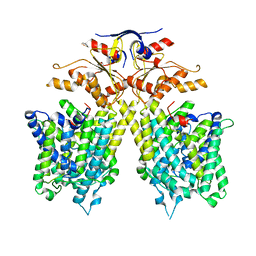 | | The overall structure of SLC26A9 | | 分子名称: | CHLORIDE ION, SODIUM ION, Solute carrier family 26 member 9 | | 著者 | Chi, X.M, Chen, Y, Li, X.R, Zhang, Y.Y, Zhou, Q. | | 登録日 | 2020-07-03 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural insights into the gating mechanism of human SLC26A9 mediated by its C-terminal sequence.
Cell Discov, 6, 2020
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | 分子名称: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | 著者 | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | 登録日 | 2020-06-01 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7C8K
 
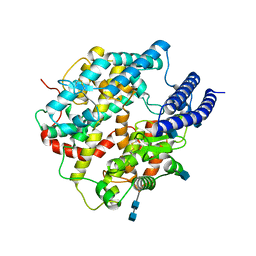 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | 登録日 | 2020-06-02 | | 公開日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7V2Z
 
 | |
3MTL
 
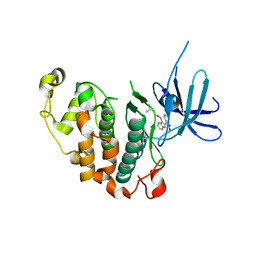 | | Crystal structure of the PCTAIRE1 kinase in complex with Indirubin E804 | | 分子名称: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cell division protein kinase 16 | | 著者 | Krojer, T, Sharpe, T.D, Roos, A, Savitsky, P, Amos, A, Ayinampudi, V, Berridge, G, Fedorov, O, Keates, T, Phillips, C, Burgess-Brown, N, Zhang, Y, Pike, A.C.W, Muniz, J, Vollmar, M, Thangaratnarajah, C, Rellos, P, Ugochukwu, E, Filippakopoulos, P, Yue, W, Das, S, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2010-04-30 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16.
Biochem.J., 474, 2017
|
|
8HIC
 
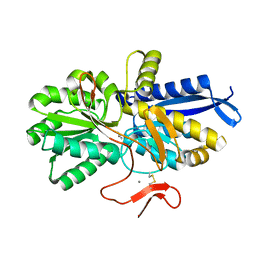 | | Crystal structure of UrtA from Prochlorococcus marinus str. MIT 9313 in complex with urea and calcium | | 分子名称: | CALCIUM ION, Putative urea ABC transporter, substrate binding protein, ... | | 著者 | Zhang, Y.Z, Wang, P, Wang, C. | | 登録日 | 2022-11-19 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and molecular basis for urea recognition by Prochlorococcus.
J.Biol.Chem., 299, 2023
|
|
7XXB
 
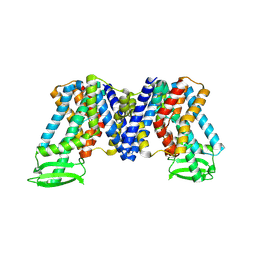 | | IAA bound state of AtPIN3 | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 3 | | 著者 | Su, N. | | 登録日 | 2022-05-29 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
8D1E
 
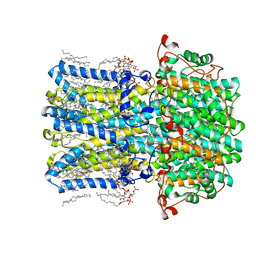 | | hBest2 1uM Ca2+ (Ca2+-bound) closed state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | 著者 | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | 登録日 | 2022-05-27 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (1.78 Å) | | 主引用文献 | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1J
 
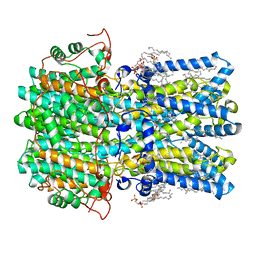 | | hBest1 5mM Ca2+ (Ca2+-bound) closed state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | 著者 | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | 登録日 | 2022-05-27 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.05 Å) | | 主引用文献 | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1L
 
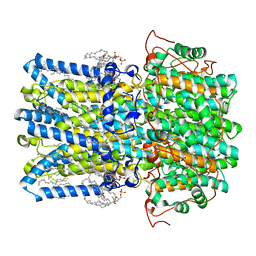 | | hBest1 Ca2+-bound partially open aperture state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION, ... | | 著者 | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | 登録日 | 2022-05-27 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.12 Å) | | 主引用文献 | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1M
 
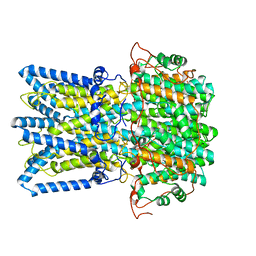 | |
8D1H
 
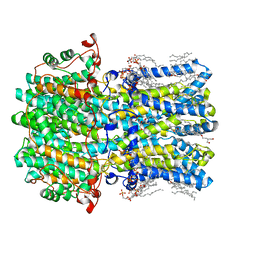 | | hBest2 Ca2+-unbound closed state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | 著者 | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | 登録日 | 2022-05-27 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (1.94 Å) | | 主引用文献 | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
6CSX
 
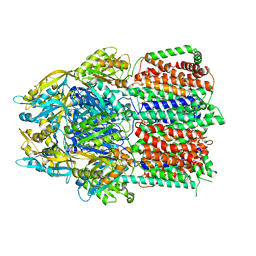 | |
7UJD
 
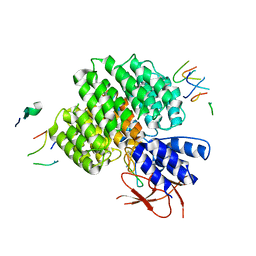 | | PSMD2 Structure bound to MC1 and Fab8/14 | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 2, ACY-PHE-PRO-ASP-VAL-SAR-LEU-HIS-ARG-TYR-TRP-GLY-TRP-ASP-CYS-GLY-NH2, Fab 14 HC CDRs, ... | | 著者 | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | 登録日 | 2022-03-30 | | 公開日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
7UIH
 
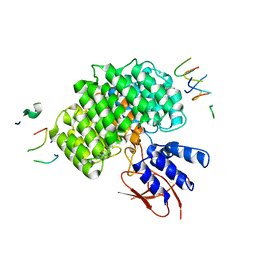 | | PSMD2 Structure | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 2, Fab 14 HC CDRs, Fab 14 LC CDRs, ... | | 著者 | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | 登録日 | 2022-03-29 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
5EJ2
 
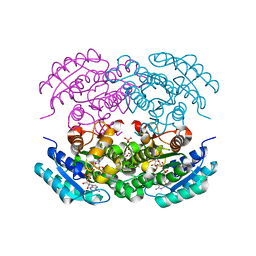 | |
5H21
 
 | |
6BX3
 
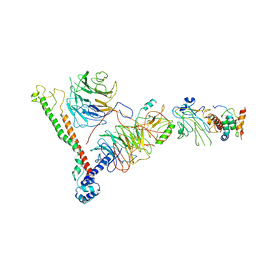 | | Structure of histone H3k4 methyltransferase | | 分子名称: | COMPASS component BRE2, COMPASS component SDC1, COMPASS component SPP1, ... | | 著者 | Skiniotis, G, Qu, Q.H. | | 登録日 | 2017-12-16 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex.
Cell, 174, 2018
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
3LLS
 
 | |
