5XVJ
 
 | | Crystal structure of AL7 PAL domain | | 分子名称: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | 著者 | Peng, L, Wang, L.L, Huang, Y. | | 登録日 | 2017-06-28 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
1CHZ
 
 | | A NEW NEUROTOXIN FROM BUTHUS MARTENSII KARSCH | | 分子名称: | CHLORIDE ION, PROTEIN (BMK M2) | | 著者 | He, X.L, Deng, J.P, Li, H.M, Wang, D.C. | | 登録日 | 1999-03-31 | | 公開日 | 2000-03-31 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structure of a new neurotoxin from the scorpion Buthus martensii Karsch at 1.76 A.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5XVM
 
 | |
5W5Q
 
 | | MAP4K4 in complex with inhibitor compound 12 (N3-methyl-10-(3-methyl-3-(5-methyloxazol-2-yl)but-1-yn-1-yl)-6,7-dihydro-5H-5,7-methanobenzo[c]imidazo[1,2-a]azepine-2,3-dicarboxamide) | | 分子名称: | (5s,7s)-N~3~-methyl-10-[3-methyl-3-(5-methyl-1,3-oxazol-2-yl)but-1-yn-1-yl]-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase kinase 4 | | 著者 | Harris, S.F, Wu, P. | | 登録日 | 2017-06-15 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Structure Based Design of Potent Selective Inhibitors of Protein Kinase D1 (PKD1).
Acs Med.Chem.Lett., 10, 2019
|
|
3C3V
 
 | |
8KIH
 
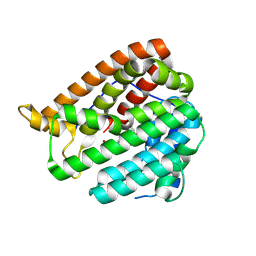 | | PhmA, a type I diterpene synthase without NST/DTE motif | | 分子名称: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, diterpene synthase, ... | | 著者 | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | 登録日 | 2023-08-23 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
8KI5
 
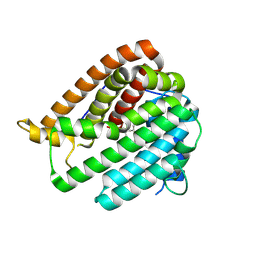 | | PhmA, a type I diterpene synthase without NST/DTE motif | | 分子名称: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, PhmA | | 著者 | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | 登録日 | 2023-08-22 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
7MQS
 
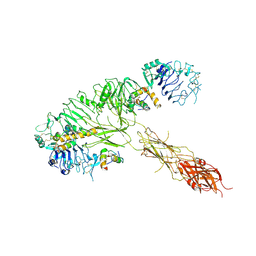 | | The insulin receptor ectodomain in complex with three venom hybrid insulin molecules - asymmetric conformation | | 分子名称: | Insulin A chain, Insulin B chain, Isoform Short of Insulin receptor | | 著者 | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | 登録日 | 2021-05-06 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
7MQO
 
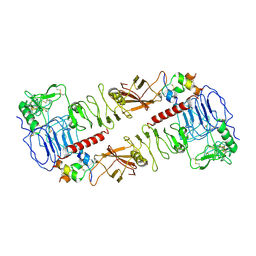 | | The insulin receptor ectodomain in complex with a venom hybrid insulin analog - "head" region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, ... | | 著者 | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | 登録日 | 2021-05-06 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
7MQR
 
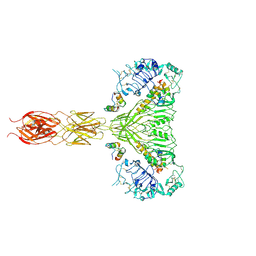 | | The insulin receptor ectodomain in complex with four venom hybrid insulins - symmetric conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | 著者 | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | 登録日 | 2021-05-06 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
6JC7
 
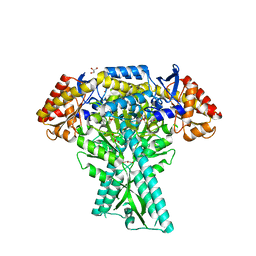 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with amino donor L-Ala | | 分子名称: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, ACETIC ACID, CrmG, ... | | 著者 | Xu, J, Su, K, Liu, J. | | 登録日 | 2019-01-28 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural studies reveal flexible roof of active site responsible for omega-transaminase CrmG overcoming by-product inhibition.
Commun Biol, 3, 2020
|
|
6JC9
 
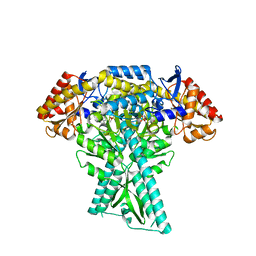 | |
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
5Y6G
 
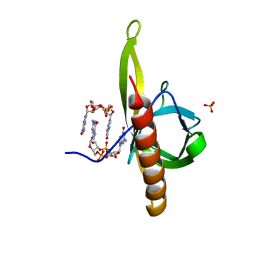 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | 著者 | Hou, Y.J, Wang, D.C, Li, D.F. | | 登録日 | 2017-08-11 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
3FBP
 
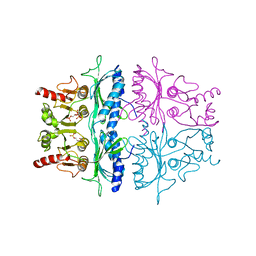 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | 著者 | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | 登録日 | 1990-06-07 | | 公開日 | 1992-04-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
5Y6H
 
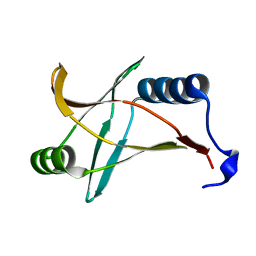 | |
6JLC
 
 | |
6JCB
 
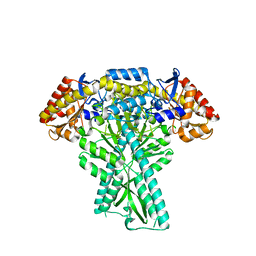 | |
5Y6F
 
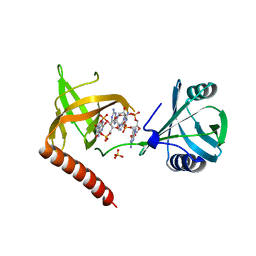 | | Crystal structure of YcgR in complex with c-di-GMP from Escherichia coli | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | 著者 | Hou, Y.J, Wang, D.C, Li, D.F. | | 登録日 | 2017-08-11 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
6JC8
 
 | |
5GYQ
 
 | |
6IEW
 
 | |
6IHJ
 
 | |
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | 著者 | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | 登録日 | 2023-12-15 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
6JIV
 
 | | SspE crystal structure | | 分子名称: | SspE protein | | 著者 | Bing, Y.Z, Yang, H.G. | | 登録日 | 2019-02-23 | | 公開日 | 2020-03-25 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
