3O05
 
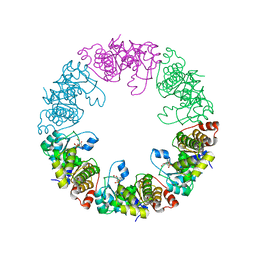 | | Crystal Structure of Yeast Pyridoxal 5-Phosphate Synthase Snz1 Complxed with Substrate PLP | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | 著者 | Teng, Y.B, Zhang, X, He, Y.X, Hu, H.X, Zhou, C.Z. | | 登録日 | 2010-07-19 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
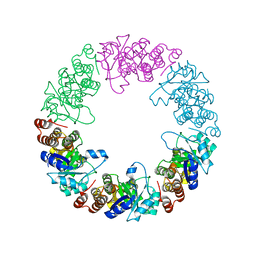
3O06
 
 | |
3O07
 
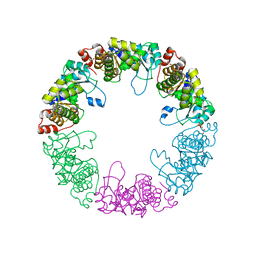 | | Crystal structure of yeast pyridoxal 5-phosphate synthase Snz1 complexed with substrate G3P | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | 著者 | Teng, Y.B, Zhang, X, Hu, H.X, Zhou, C.Z. | | 登録日 | 2010-07-19 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
7DY9
 
 | | Thermotoga maritima ferritin mutant-FLAL | | 分子名称: | Ferritin | | 著者 | Zhao, G, Zhang, X. | | 登録日 | 2021-01-20 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7DYB
 
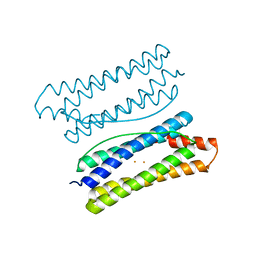 | | Thermotoga maritima ferritin mutant-FLAL-L | | 分子名称: | FE (III) ION, Ferritin | | 著者 | Zhao, G, Zhang, X. | | 登録日 | 2021-01-20 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.779 Å) | | 主引用文献 | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | 著者 | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | 登録日 | 2019-11-20 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | 分子名称: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | 著者 | Zhang, X, Ma, J, Wang, Y, Wang, J. | | 登録日 | 2016-08-07 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
7X5B
 
 | | Crystal structure of RuvB | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X5A
 
 | | CryoEM structure of RuvA-Holliday junction complex | | 分子名称: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | 分子名称: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | 分子名称: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
1RHP
 
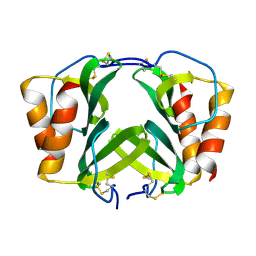 | |
8XJM
 
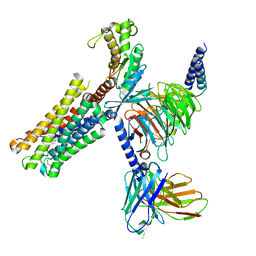 | | Latanoprost acid bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | 分子名称: | Antibody fragment scFv16, Engineered miniGq, Fusion tag,Prostaglandin F2-alpha receptor,LgBiT, ... | | 著者 | Zhang, X, Li, X, Liu, G, Gong, W. | | 登録日 | 2023-12-21 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJK
 
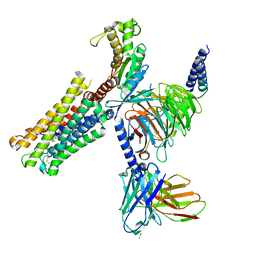 | | Cloprosetnol bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | 分子名称: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | 著者 | Zhang, X, Li, X, Liu, G, Gong, W. | | 登録日 | 2023-12-21 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJO
 
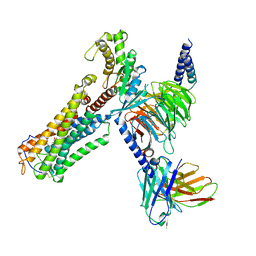 | | U46619 bound Thromboxane A2 receptor-Gq Protein Complex | | 分子名称: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | 著者 | Zhang, X, Li, X, Liu, G, Gong, W. | | 登録日 | 2023-12-21 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJN
 
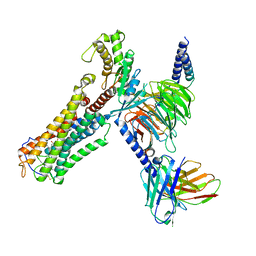 | | Cloprosetnol bound Thromboxane A2 receptor-Gq Protein Complex | | 分子名称: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | 著者 | Zhang, X, Li, X, Liu, G, Gong, W. | | 登録日 | 2023-12-21 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJL
 
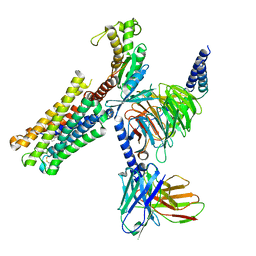 | | PGF2-alpha bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | 分子名称: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | 著者 | Zhang, X, Li, X, Liu, G, Gong, W. | | 登録日 | 2023-12-21 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
5T7Q
 
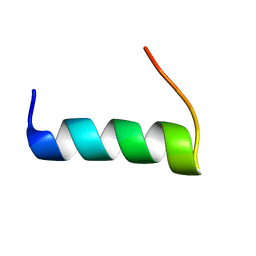 | | TIRAP phosphoinositide-binding motif | | 分子名称: | Toll/interleukin-1 receptor domain-containing adapter protein | | 著者 | Capelluto, D.G.S, Ellena, J.F, Armstrong, G, Zhao, X, Xiao, S. | | 登録日 | 2016-09-05 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Membrane targeting of TIRAP is negatively regulated by phosphorylation in its phosphoinositide-binding motif.
Sci Rep, 7, 2017
|
|
4O9L
 
 | |
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | 分子名称: | D-Cysteine desulfhydrase | | 著者 | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-08-12 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | 分子名称: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | 著者 | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-08-12 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
4O9F
 
 | |
3T9N
 
 | | Crystal structure of a membrane protein | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | 著者 | Yang, M, Zhang, X, Ge, J, Wang, J. | | 登録日 | 2011-08-03 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.456 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4RSQ
 
 | | 2.9A resolution structure of SRPN2 (K198C/E359C) from Anopheles gambiae | | 分子名称: | Serpin 2 | | 著者 | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | 登録日 | 2014-11-10 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
4ROA
 
 | | 1.90A resolution structure of SRPN2 (S358W) from Anopheles gambiae | | 分子名称: | Serpin 2 | | 著者 | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | 登録日 | 2014-10-28 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
