5W3O
 
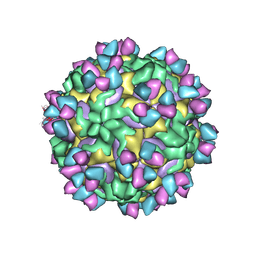 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, empty particle) | | 分子名称: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | 著者 | Liu, Y, Dong, Y, Rossmann, M.G. | | 登録日 | 2017-06-08 | | 公開日 | 2017-07-12 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3L
 
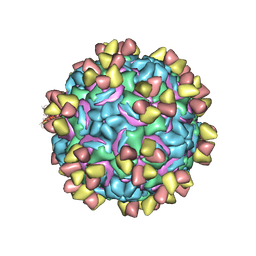 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degrees Celsius, molar ratio 1:3, full particle) | | 分子名称: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | 著者 | Liu, Y, Dong, Y, Rossmann, M.G. | | 登録日 | 2017-06-08 | | 公開日 | 2017-07-12 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3E
 
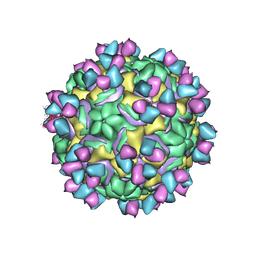 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, full particle) | | 分子名称: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | 著者 | Liu, Y, Dong, Y, Rossmann, M.G. | | 登録日 | 2017-06-07 | | 公開日 | 2017-07-12 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ASG
 
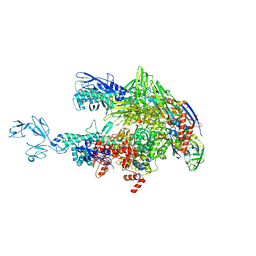 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | 登録日 | 2017-08-24 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
4EQK
 
 | |
4ENF
 
 | | Crystal structure of the cap-binding domain of polymerase basic protein 2 from influenza virus A/Puerto Rico/8/34(h1n1) | | 分子名称: | 1,4-BUTANEDIOL, NITRATE ION, Polymerase basic protein 2 | | 著者 | Meng, G, Liu, Y, Zheng, X. | | 登録日 | 2012-04-13 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus
J.Biol.Chem., 288, 2013
|
|
4ES5
 
 | |
3G3M
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-iodo-UMP | | 分子名称: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | 著者 | Liu, Y, Tang, H.L, Bello, A.M, Poduch, E, Kotra, L.P, Pai, E.F. | | 登録日 | 2009-02-02 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
3G3D
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-azido-UMP | | 分子名称: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | 著者 | Liu, Y, Tang, H.L, Bello, A, Poduch, E, Kotra, L, Pai, E. | | 登録日 | 2009-02-02 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
4PLI
 
 | |
4PLL
 
 | |
3MI2
 
 | | Crystal structure of human orotidine-5'-monophosphate decarboxylase complexed with pyrazofurin monophosphate | | 分子名称: | (1S)-1,4-anhydro-1-(5-carbamoyl-4-hydroxy-1H-pyrazol-3-yl)-5-O-phosphono-D-ribitol, Uridine 5'-monophosphate synthase | | 著者 | Liu, Y, To, T, Kotra, L.P, Pai, E.F. | | 登録日 | 2010-04-09 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural determinants for the inhibitory ligands of orotidine-5'-monophosphate decarboxylase.
Bioorg.Med.Chem., 18, 2010
|
|
3MO7
 
 | |
4RGY
 
 | |
8CVY
 
 | |
8CVX
 
 | | Human glycogenin-1 and glycogen synthase-1 complex in the presence of glucose-6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, Glycogen [starch] synthase, muscle, ... | | 著者 | Liu, Y, Fastman, N.M, Tzitzilonis, C. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The structural mechanism of human glycogen synthesis by the GYS1-GYG1 complex.
Cell Rep, 40, 2022
|
|
8CVZ
 
 | |
5H06
 
 | | Crystal structure of AmyP in complex with maltose | | 分子名称: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | He, C, Liu, Y. | | 登録日 | 2016-10-03 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
5H05
 
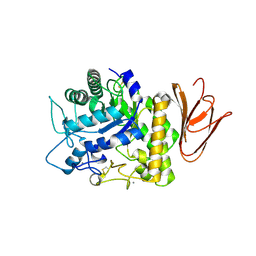 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | 分子名称: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | He, C, Liu, Y. | | 登録日 | 2016-10-03 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
4ZK7
 
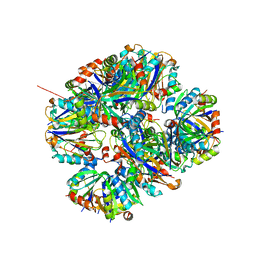 | | Crystal structure of rescued two-component self-assembling tetrahedral cage T33-31 | | 分子名称: | Chorismate mutase, Divalent-cation tolerance protein CutA | | 著者 | Liu, Y, Cascio, D, Sawaya, M.R, Bale, J, Collazo, M.J, Park, R, King, N, Baker, D, Yeates, T. | | 登録日 | 2015-04-30 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci., 24, 2015
|
|
4PEJ
 
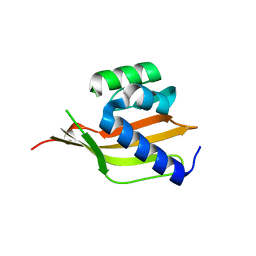 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | 分子名称: | Retro-aldolase | | 著者 | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | 登録日 | 2014-04-23 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEK
 
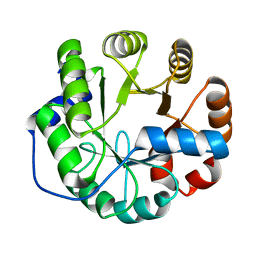 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | 分子名称: | Retro-aldolase | | 著者 | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | 登録日 | 2014-04-23 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
6IH4
 
 | |
6IH5
 
 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | 分子名称: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | 登録日 | 2018-09-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.468 Å) | | 主引用文献 | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH8
 
 | |
