7WJR
 
 | |
1UA3
 
 | |
1VXA
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXD
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXG
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXF
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXH
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXC
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXB
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXE
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1GFL
 
 | |
3EZM
 
 | | CYANOVIRIN-N | | Descriptor: | PROTEIN (CYANOVIRIN-N) | | Authors: | Yang, F, Wlodawer, A. | | Deposit date: | 1998-12-15 | | Release date: | 1998-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping.
J.Mol.Biol., 288, 1999
|
|
3P4L
 
 | |
1SPE
 
 | | SPERM WHALE NATIVE CO MYOGLOBIN AT PH 4.0, TEMP 4C | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
5T3Y
 
 | |
1BG8
 
 | |
1C5E
 
 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
4ZEF
 
 | |
4G4P
 
 | | Crystal structure of glutamine-binding protein from Enterococcus faecalis at 1.5 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Amino acid ABC transporter, amino acid-binding/permease protein, ... | | Authors: | Fulyani, F, Guskov, A, Zagar, A.V, Slotboom, D.-J, Poolman, B. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
5GIY
 
 | | HSA-Palmitic acid-[RuCl5(ind)]2- | | Descriptor: | PALMITIC ACID, Serum albumin, pentakis(chloranyl)-(1~{H}-indazol-2-ium-2-yl)ruthenium(1-) | | Authors: | Yang, F, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structure of HSA-Palmitic acid-[RuCl5(ind)]2-
To Be Published
|
|
1OM0
 
 | | crystal structure of xylanase inhibitor protein (XIP-I) from wheat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Xylanase Inhibitor Protein I | | Authors: | Payan, F, Flatman, R, Porciero, S, Williamson, G, Juge, N, Roussel, A. | | Deposit date: | 2003-02-24 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson).
Biochem.J., 372, 2003
|
|
1TA3
 
 | | Crystal Structure of xylanase (GH10) in complex with inhibitor (XIP) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
1TE1
 
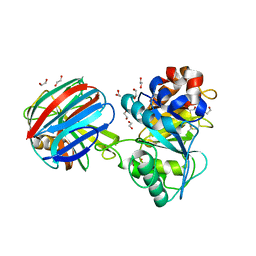 | | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, endo-1,4-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-24 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
2I8L
 
 | |
2NAX
 
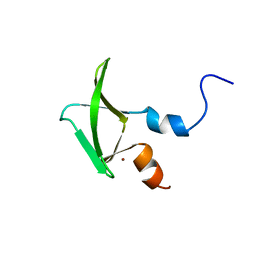 | | Structure of CCHC zinc finger domain of Pcf11 | | Descriptor: | Protein PCF11, ZINC ION | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C terminus of Pcf11 forms a novel zinc-finger structure that plays an essential role in mRNA 3'-end processing.
RNA, 23, 2017
|
|
