3DSC
 
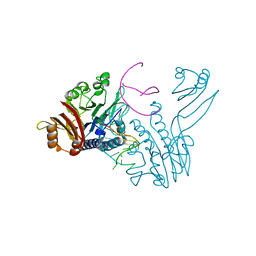 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | Descriptor: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | Authors: | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3DSD
 
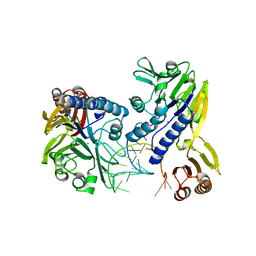 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | Authors: | Williams, R.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3HUE
 
 | |
3HUF
 
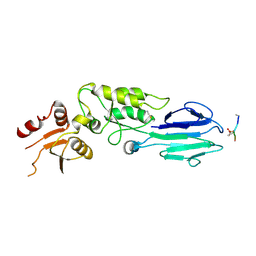 | | Structure of the S. pombe Nbs1-Ctp1 complex | | Descriptor: | DNA repair and telomere maintenance protein nbs1, Double-strand break repair protein ctp1, THIOCYANATE ION | | Authors: | Williams, R.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-06-13 | | Release date: | 2009-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair.
Cell(Cambridge,Mass.), 139, 2009
|
|
1JNX
 
 | |
1N5O
 
 | |
1T2V
 
 | | Structural basis of phospho-peptide recognition by the BRCT domain of BRCA1, structure with phosphopeptide | | Descriptor: | BRCTide-7PS, Breast cancer type 1 susceptibility protein | | Authors: | Williams, R.S, Lee, M.S, Hau, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T2U
 
 | | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1: structure of BRCA1 missense variant V1809F | | Descriptor: | Breast cancer type 1 susceptibility protein, COBALT (II) ION, SULFATE ION | | Authors: | Williams, R.S, Lee, M.S, Duong, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
8FTK
 
 | | Chaetomium thermophilum SETX (Full-length) | | Descriptor: | 5'-3' RNA helicase-like protein | | Authors: | Williams, R.S, Appel, C.D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | Sen1 architecture: RNA-DNA hybrid resolution, autoregulation, and insights into SETX inactivation in AOA2.
Mol.Cell, 83, 2023
|
|
8FTM
 
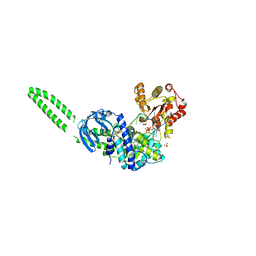 | | Setx-ssRNA-ADP-SO4 complex | | Descriptor: | 5'-3' RNA helicase-like protein, ADENOSINE-5'-DIPHOSPHATE, RNA (5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Williams, R.S, Appel, C.D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Sen1 architecture: RNA-DNA hybrid resolution, autoregulation, and insights into SETX inactivation in AOA2.
Mol.Cell, 83, 2023
|
|
8FTH
 
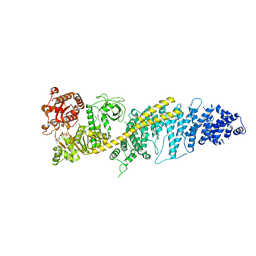 | | Chaetomium thermophilum SETX - NPPC internal deletion | | Descriptor: | 5'-3' RNA helicase-like protein, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Williams, R.S, Appel, C.D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Sen1 architecture: RNA-DNA hybrid resolution, autoregulation, and insights into SETX inactivation in AOA2.
Mol.Cell, 83, 2023
|
|
3L41
 
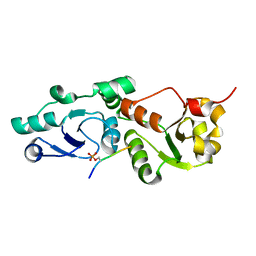 | | Crystal Structure of S. pombe Brc1 BRCT5-BRCT6 domains in complex with phosphorylated H2A | | Descriptor: | BRCT-containing protein 1, phosphorylated H2A tail | | Authors: | Williams, R.S, Williams, J.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | gammaH2A binds Brc1 to maintain genome integrity during S-phase.
Embo J., 29, 2010
|
|
3L40
 
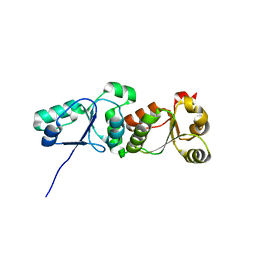 | |
5KIV
 
 | | Crystal structure of SauMacro (SAV0325) | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Protein-ADP-ribose hydrolase, ... | | Authors: | Williams, R.S, Appel, C.D, Feld, G.K, Wallace, B.D. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the sirtuin-linked macrodomain SAV0325 from Staphylococcus aureus.
Protein Sci., 25, 2016
|
|
7UOA
 
 | | MAGEA4-MTP1 linear peptide complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MTP-1, ... | | Authors: | Williams, R.S, Tumbale, P.S. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery and Structural Basis of the Selectivity of Potent Cyclic Peptide Inhibitors of MAGE-A4.
J.Med.Chem., 65, 2022
|
|
5INK
 
 | |
5INM
 
 | |
3SZQ
 
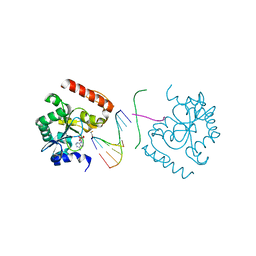 | | Structure of an S. pombe APTX/DNA/AMP/Zn complex | | Descriptor: | 5'-D(*CP*CP*CP*TP*G)-3', 5'-D(*TP*AP*TP*CP*GP*GP*AP*AP*TP*CP*AP*GP*GP*G)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Tumbale, P, Krahn, J, Williams, R.S. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7N3Z
 
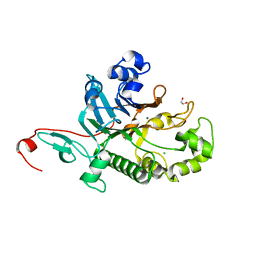 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
7N3Y
 
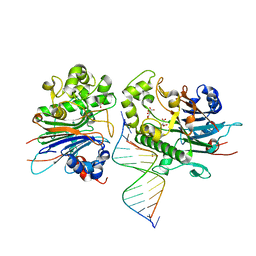 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant in Complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Wojtaszek, J.L, Krahn, J, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
1YJ5
 
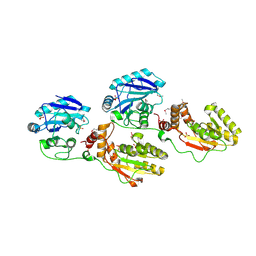 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1YJM
 
 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
4NCI
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation | | Descriptor: | DNA double-strand break repair Rad50 ATPase | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCJ
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation with ADP Beryllium Flouride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA double-strand break repair Rad50 ATPase, ... | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCK
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation | | Descriptor: | CHLORIDE ION, DNA double-strand break repair Rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
