6WPS
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6WS6
 
 | | Structural and functional analysis of a potent sarbecovirus neutralizing antibody | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), S309 antigen-binding (Fab) fragment, heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, Marco, A.D, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Telenti, A, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6WPT
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
2ABO
 
 | | NMR structure of gamma herpesvirus 68 a viral Bcl-2 homolog | | Descriptor: | bcl-2 homolog | | Authors: | Loh, J, Huang, Q, Petros, A.M, Nettesheim, D, van Dyk, L.F, Labrada, L, Speck, S.H, Levine, B, Olejniczak, E.T, Virgin, H.W. | | Deposit date: | 2005-07-15 | | Release date: | 2006-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A surface groove essential for viral Bcl-2 function during chronic infection in vivo.
Plos Pathog., 1, 2005
|
|
1K3K
 
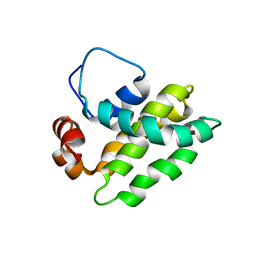 | | Solution Structure of a Bcl-2 Homolog from Kaposi's Sarcoma Virus | | Descriptor: | functional anti-apoptotic factor vBCL-2 homolog | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q59
 
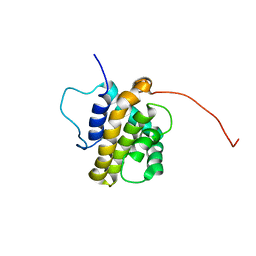 | | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2 | | Descriptor: | Early antigen protein R | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2
J.Mol.Biol., 332, 2003
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6CRJ
 
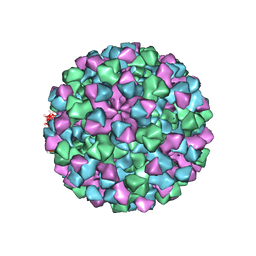 | |
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6P4L
 
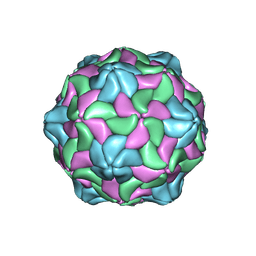 | |
6P4J
 
 | | Mouse norovirus complexed with GCDCA | | Descriptor: | Capsid protein, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Smith, T.J, Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
6P4K
 
 | | mouse norovirus complexed with TCA | | Descriptor: | Capsid protein, TAUROCHOLIC ACID | | Authors: | Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
5FFL
 
 | |
6E47
 
 | |
6E48
 
 | | Crystal Structure of the Murine Norovirus VP1 P domain in complex with the CD300lf Receptor and Lithocholic Acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8ERQ
 
 | |
8ERR
 
 | |
7N8I
 
 | |
7N8H
 
 | |
8G30
 
 | |
1ML0
 
 | |
1MKF
 
 | |
6C6Q
 
 | |
6C74
 
 | |
7TAS
 
 | |
