1NZO
 
 | | The crystal structure of wild type penicillin-binding protein 5 from E. coli | | Descriptor: | BETA-MERCAPTOETHANOL, Penicillin-binding protein 5 | | Authors: | Nicholas, R.A, Krings, S, Tomberg, J, Nicola, G, Davies, C. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of wild-type penicillin-binding protein 5 from Escherichia coli: implications for deacylation of the acyl-enzyme complex.
J.Biol.Chem., 278, 2003
|
|
1NJ4
 
 | |
1SDN
 
 | |
3MZD
 
 | | Structure of penicillin-binding protein 5 from E. coli: cloxacillin acyl-enzyme complex | | Descriptor: | (2R,4S)-2-[(1S)-1-({[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}amino)-2-oxoethyl]-5,5-dimethyl-1,3-thiazolid ine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3MZE
 
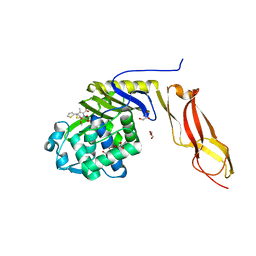 | | Structure of penicillin-binding protein 5 from E.coli: cefoxitin acyl-enzyme complex | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3MZF
 
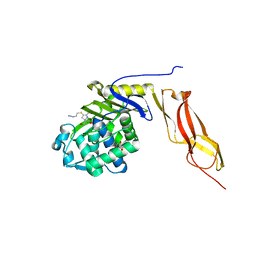 | | Structure of penicillin-binding protein 5 from E. coli: imipenem acyl-enzyme complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
1HD8
 
 | |
1NZU
 
 | |
1Z6F
 
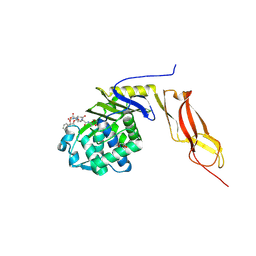 | | Crystal structure of penicillin-binding protein 5 from E. coli in complex with a boronic acid inhibitor | | Descriptor: | GLYCEROL, N1-[(1R)-1-(DIHYDROXYBORYL)ETHYL]-N2-[(TERT-BUTOXYCARBONYL)-D-GAMMA-GLUTAMYL]-N6-[(BENZYLOXY)CARBONYL-L-LYSINAMIDE, Penicillin-binding protein 5 | | Authors: | Nicola, G, Peddi, S, Stefanova, M, Nicholas, R.A, Gutheil, W.G, Davies, C. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Escherichia coli Penicillin-Binding Protein 5 Bound to a Tripeptide Boronic Acid Inhibitor: A Role for Ser-110 in Deacylation.
Biochemistry, 44, 2005
|
|
3EQV
 
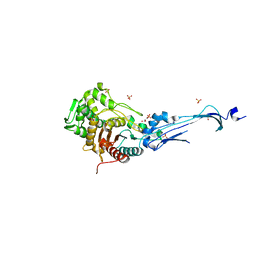 | | Crystal structure of penicillin-binding protein 2 from Neisseria gonorrhoeae containing four mutations associated with penicillin resistance | | Descriptor: | GLYCEROL, Penicillin-binding protein 2, SULFATE ION | | Authors: | Powell, A.J, Deacon, A.M, Nicholas, R.A, Davies, C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-binding Protein 2 from Penicillin-susceptible and -resistant Strains of Neisseria gonorrhoeae Reveal an Unexpectedly Subtle Mechanism for Antibiotic Resistance.
J.Biol.Chem., 284, 2009
|
|
3EQU
 
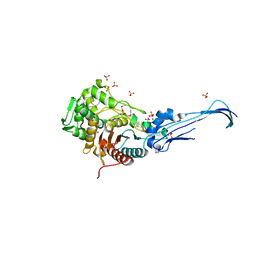 | | Crystal structure of penicillin-binding protein 2 from Neisseria gonorrhoeae | | Descriptor: | GLYCEROL, Penicillin-binding protein 2, SULFATE ION | | Authors: | Powell, A.J, Deacon, A.M, Nicholas, R.A, Davies, C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-binding Protein 2 from Penicillin-susceptible and -resistant Strains of Neisseria gonorrhoeae Reveal an Unexpectedly Subtle Mechanism for Antibiotic Resistance.
J.Biol.Chem., 284, 2009
|
|
2G6G
 
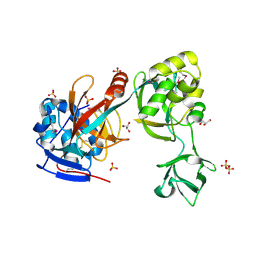 | | Crystal structure of MltA from Neisseria gonorrhoeae | | Descriptor: | GLYCEROL, GNA33, SULFATE ION | | Authors: | Powell, A.J, Liu, Z.J, Nicholas, R.A, Davies, C. | | Deposit date: | 2006-02-24 | | Release date: | 2006-05-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the Lytic Transglycosylase MltA from N.gonorrhoeae and E.coli: Insights into Interdomain Movements and Substrate Binding.
J.Mol.Biol., 359, 2006
|
|
2GAE
 
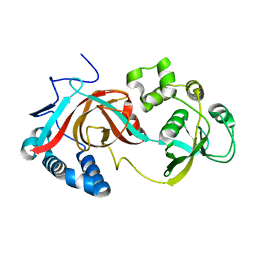 | | Crystal structure of MltA from E. coli | | Descriptor: | Membrane-bound lytic murein transglycosylase A | | Authors: | Powell, A.J, Liu, Z.J, Nicholas, R.A, Davies, C. | | Deposit date: | 2006-03-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the Lytic Transglycosylase MltA from N.gonorrhoeae and E.coli: Insights into Interdomain Movements and Substrate Binding.
J.Mol.Biol., 359, 2006
|
|
2GK2
 
 | | Crystal structure of the N terminal domain of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, GLYCEROL, NICKEL (II) ION | | Authors: | Fedarovich, A, Tomberg, J, Nicholas, R.A, Davies, C. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the N-terminal domain of human CEACAM1: binding target of the opacity proteins during invasion of Neisseria meningitidis and N. gonorrhoeae.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3LO7
 
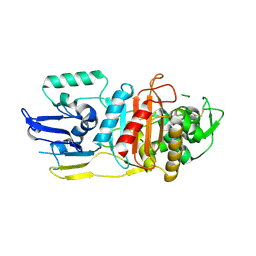 | |
3UPN
 
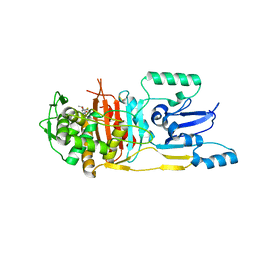 | |
3UPP
 
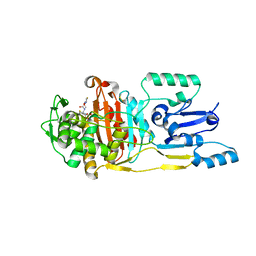 | |
3UPO
 
 | |
3UN7
 
 | |
6XQV
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | Descriptor: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQZ
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQY
 
 | |
6XQX
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
5KSH
 
 | |
4U3T
 
 | |
